The metazoan Mediator co-activator complex as an integrative hub for transcriptional regulation
- PMID: 20940737
- PMCID: PMC3217725
- DOI: 10.1038/nrg2901
The metazoan Mediator co-activator complex as an integrative hub for transcriptional regulation
Abstract
The Mediator is an evolutionarily conserved, multiprotein complex that is a key regulator of protein-coding genes. In metazoan cells, multiple pathways that are responsible for homeostasis, cell growth and differentiation converge on the Mediator through transcriptional activators and repressors that target one or more of the almost 30 subunits of this complex. Besides interacting directly with RNA polymerase II, Mediator has multiple functions and can interact with and coordinate the action of numerous other co-activators and co-repressors, including those acting at the level of chromatin. These interactions ultimately allow the Mediator to deliver outputs that range from maximal activation of genes to modulation of basal transcription to long-term epigenetic silencing.
Conflict of interest statement
The authors declare no competing financial interests.
Figures

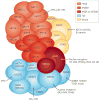


References
-
- Roeder RG. Transcriptional regulation and the role of diverse coactivators in animal cells. FEBS Lett. 2005;579:909–915. - PubMed
-
- Kornberg RD. Mediator and the mechanism of transcriptional activation. Trends Biochem Sci. 2005;30:235–239. - PubMed
-
- Malik S, Roeder RG. Dynamic regulation of Pol II transcription by the mammalian Mediator complex. Trends Biochem Sci. 2005;30:256–263. - PubMed
-
- Malik S, Roeder RG. Transcriptional regulation through Mediator-like coactivators in yeast and metazoan cells. Trends Biochem Sci. 2000;25:277–283. - PubMed
-
- Lee TI, Young RA. Transcription of eukaryotic protein-coding genes. Annu Rev Genet. 2000;34:77–137. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

