Molecular and evolutionary bases of within-patient genotypic and phenotypic diversity in Escherichia coli extraintestinal infections
- PMID: 20941353
- PMCID: PMC2947995
- DOI: 10.1371/journal.ppat.1001125
Molecular and evolutionary bases of within-patient genotypic and phenotypic diversity in Escherichia coli extraintestinal infections
Erratum in
- PLoS Pathog. 2011 Jun;7(6). doi:10.1371/annotation/700dcb3b-3475-49a1-9064-d9d070cda7be. Cruvellier, Stéphane[corrected to Cruveiller, Stéphane]
Abstract
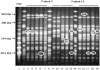
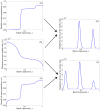
Although polymicrobial infections, caused by combinations of viruses, bacteria, fungi and parasites, are being recognised with increasing frequency, little is known about the occurrence of within-species diversity in bacterial infections and the molecular and evolutionary bases of this diversity. We used multiple approaches to study the genomic and phenotypic diversity among 226 Escherichia coli isolates from deep and closed visceral infections occurring in 19 patients. We observed genomic variability among isolates from the same site within 11 patients. This diversity was of two types, as patients were infected either by several distinct E. coli clones (4 patients) or by members of a single clone that exhibit micro-heterogeneity (11 patients); both types of diversity were present in 4 patients. A surprisingly wide continuum of antibiotic resistance, outer membrane permeability, growth rate, stress resistance, red dry and rough morphotype characteristics and virulence properties were present within the isolates of single clones in 8 of the 11 patients showing genomic micro-heterogeneity. Many of the observed phenotypic differences within clones affected the trade-off between self-preservation and nutritional competence (SPANC). We showed in 3 patients that this phenotypic variability was associated with distinct levels of RpoS in co-existing isolates. Genome mutational analysis and global proteomic comparisons in isolates from a patient revealed a star-like relationship of changes amongst clonally diverging isolates. A mathematical model demonstrated that multiple genotypes with distinct RpoS levels can co-exist as a result of the SPANC trade-off. In the cases involving infection by a single clone, we present several lines of evidence to suggest diversification during the infectious process rather than an infection by multiple isolates exhibiting a micro-heterogeneity. Our results suggest that bacteria are subject to trade-offs during an infectious process and that the observed diversity resembled results obtained in experimental evolution studies. Whatever the mechanisms leading to diversity, our results have strong medical implications in terms of the need for more extensive isolate testing before deciding on antibiotic therapies.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures





References
-
- Romling U, Fiedler B, Bosshammer J, Grothues D, Greipel J, et al. Epidemiology of chronic Pseudomonas aeruginosa infections in cystic fibrosis. J Infect Dis. 1994;170:1616–1621. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

