A novel approach for the generation of genetically modified mammary epithelial cell cultures yields new insights into TGFβ signaling in the mammary gland
- PMID: 20942910
- PMCID: PMC3096976
- DOI: 10.1186/bcr2728
A novel approach for the generation of genetically modified mammary epithelial cell cultures yields new insights into TGFβ signaling in the mammary gland
Abstract
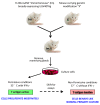
Introduction: Molecular dissection of the signaling pathways that underlie complex biological responses in the mammary epithelium is limited by the difficulty of propagating large numbers of mouse mammary epithelial cells, and by the inability of ribonucleic acid interference (RNAi)-based knockdown approaches to fully ablate gene function. Here we describe a method for the generation of conditionally immortalized mammary epithelial cells with defined genetic defects, and we show how such cells can be used to investigate complex signal transduction processes using the transforming growth factor beta (TGFβ/Smad pathway as an example.
Methods: We intercrossed the previously described H-2Kb-tsA58 transgenic mouse (Immortomouse) which expresses a temperature-sensitive mutant of the simian virus-40 large T-antigen (tsTAg), with mice of differing Smad genotypes. A panel of conditionally immortalized mammary epithelial cell (IMEC) cultures were derived from the virgin mammary glands of offspring of these crosses and used to assess the Smad dependency of different biological responses to TGFβ.
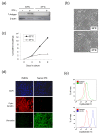
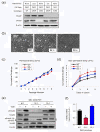
Results: IMECs could be propagated indefinitely at permissive temperatures and had a stable epithelial phenotype, resembling primary mammary epithelial cells with respect to several criteria, including responsiveness to TGFβ. Using this panel of cells, we demonstrated that Smad3, but not Smad2, is necessary for TGFβ-induced apoptotic, growth inhibitory and EMT responses, whereas either Smad can support TGFβ-induced invasion as long as a threshold level of total Smad is exceeded.
Conclusions: This work demonstrates the practicality and utility of generating conditionally immortalized mammary epithelial cell lines from genetically modified Immortomice for detailed investigation of complex signaling pathways in the mammary epithelium.
Figures




Similar articles
-
Smad3 is a key nonredundant mediator of transforming growth factor beta signaling in Nme mouse mammary epithelial cells.Mol Cancer Res. 2009 Aug;7(8):1342-53. doi: 10.1158/1541-7786.MCR-08-0558. Epub 2009 Aug 11. Mol Cancer Res. 2009. PMID: 19671686
-
A positive role for Myc in TGFbeta-induced Snail transcription and epithelial-to-mesenchymal transition.Oncogene. 2009 Jan 22;28(3):422-30. doi: 10.1038/onc.2008.395. Epub 2008 Nov 3. Oncogene. 2009. PMID: 18978814
-
ShcA Protects against Epithelial-Mesenchymal Transition through Compartmentalized Inhibition of TGF-β-Induced Smad Activation.PLoS Biol. 2015 Dec 17;13(12):e1002325. doi: 10.1371/journal.pbio.1002325. eCollection 2015 Dec. PLoS Biol. 2015. PMID: 26680585 Free PMC article.
-
A tale of two proteins: differential roles and regulation of Smad2 and Smad3 in TGF-beta signaling.J Cell Biochem. 2007 May 1;101(1):9-33. doi: 10.1002/jcb.21255. J Cell Biochem. 2007. PMID: 17340614 Review.
-
TGFβ signaling and congenital heart disease: Insights from mouse studies.Birth Defects Res A Clin Mol Teratol. 2011 Jun;91(6):423-34. doi: 10.1002/bdra.20794. Epub 2011 Apr 28. Birth Defects Res A Clin Mol Teratol. 2011. PMID: 21538815 Review.
Cited by
-
Effects on human transcriptome of mutated BRCA1 BRCT domain: a microarray study.BMC Cancer. 2012 May 30;12:207. doi: 10.1186/1471-2407-12-207. BMC Cancer. 2012. PMID: 22646717 Free PMC article.
-
Carcinogen 7,12-dimethylbenz[a]anthracene-induced mammary tumorigenesis is accelerated in Smad3 heterozygous mice compared to Smad3 wild type mice.Oncotarget. 2016 Oct 4;7(40):64878-64885. doi: 10.18632/oncotarget.11713. Oncotarget. 2016. PMID: 27588495 Free PMC article.
-
Establishment of novel in vitro mouse chief cell and SPEM cultures identifies MAL2 as a marker of metaplasia in the stomach.Am J Physiol Gastrointest Liver Physiol. 2014 Oct 15;307(8):G777-92. doi: 10.1152/ajpgi.00169.2014. Epub 2014 Sep 4. Am J Physiol Gastrointest Liver Physiol. 2014. PMID: 25190476 Free PMC article.
-
Release the ink4a/arf growth suppression by "u" and "me"?Cell Cycle. 2011 Jan 15;10(2):185-6. doi: 10.4161/cc.10.2.14471. Epub 2011 Jan 15. Cell Cycle. 2011. PMID: 21239875 Free PMC article. No abstract available.
-
Curcumin suppresses intestinal microvascular endothelial cells invasion and angiogenesis induced by activated platelets.Exp Ther Med. 2019 Aug;18(2):1099-1106. doi: 10.3892/etm.2019.7662. Epub 2019 Jun 11. Exp Ther Med. 2019. PMID: 31316605 Free PMC article.
References
-
- Derynck R, Miyazono K. The TGF-β Family. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2008. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

