Confronting two-pair primer design for enzyme-free SNP genotyping based on a genetic algorithm
- PMID: 20942913
- PMCID: PMC2964683
- DOI: 10.1186/1471-2105-11-509
Confronting two-pair primer design for enzyme-free SNP genotyping based on a genetic algorithm
Abstract
Background: Polymerase chain reaction with confronting two-pair primers (PCR-CTPP) method produces allele-specific DNA bands of different lengths by adding four designed primers and it achieves the single nucleotide polymorphism (SNP) genotyping by electrophoresis without further steps. It is a time- and cost-effective SNP genotyping method that has the advantage of simplicity. However, computation of feasible CTPP primers is still challenging.
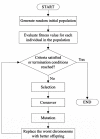
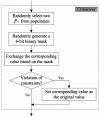
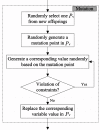
Results: In this study, we propose a GA (genetic algorithm)-based method to design a feasible CTPP primer set to perform a reliable PCR experiment. The SLC6A4 gene was tested with 288 SNPs for dry dock experiments which indicated that the proposed algorithm provides CTPP primers satisfied most primer constraints. One SNP rs12449783 in the SLC6A4 gene was taken as an example for the genotyping experiments using electrophoresis which validated the GA-based design method as providing reliable CTPP primer sets for SNP genotyping.
Conclusions: The GA-based CTPP primer design method provides all forms of estimation for the common primer constraints of PCR-CTPP. The GA-CTPP program is implemented in JAVA and a user-friendly input interface is freely available at http://bio.kuas.edu.tw/ga-ctpp/.
Figures



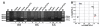
References
-
- Jasmine F, Ahsan H, Andrulis IL, John EM, Chang-Claude J, Kibriya MG. Whole-genome amplification enables accurate genotyping for microarray-based high-density single nucleotide polymorphism array. Cancer Epidemiol Biomarkers Prev. 2008;17(12):3499–3508. doi: 10.1158/1055-9965.EPI-08-0482. - DOI - PMC - PubMed
-
- Hui L, DelMonte T, Ranade K. Genotyping using the TaqMan assay. Curr Protoc Hum Genet. 2008;Chapter 2(Unit 2):10. - PubMed
-
- Yen CY, Liu SY, Chen CH, Tseng HF, Chuang LY, Yang CH, Lin YC, Wen CH, Chiang WF, Ho CH. et al.Combinational polymorphisms of four DNA repair genes XRCC1, XRCC2, XRCC3, and XRCC4 and their association with oral cancer in Taiwan. J Oral Pathol Med. 2008;37(5):271–277. doi: 10.1111/j.1600-0714.2007.00608.x. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

