Strain-specific differences in pili formation and the interaction of Corynebacterium diphtheriae with host cells
- PMID: 20942914
- PMCID: PMC2965157
- DOI: 10.1186/1471-2180-10-257
Strain-specific differences in pili formation and the interaction of Corynebacterium diphtheriae with host cells
Abstract
Background: Corynebacterium diphtheriae, the causative agent of diphtheria, is well-investigated in respect to toxin production, while little is known about C. diphtheriae factors crucial for colonization of the host. In this study, we investigated strain-specific differences in adhesion, invasion and intracellular survival and analyzed formation of pili in different isolates.
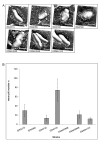
Results: Adhesion of different C. diphtheriae strains to epithelial cells and invasion of these cells are not strictly coupled processes. Using ultrastructure analyses by atomic force microscopy, significant differences in macromolecular surface structures were found between the investigated C. diphtheriae strains in respect to number and length of pili. Interestingly, adhesion and pili formation are not coupled processes and also no correlation between invasion and pili formation was found. Using RNA hybridization and Western blotting experiments, strain-specific pili expression patterns were observed. None of the studied C. diphtheriae strains had a dramatic detrimental effect on host cell viability as indicated by measurements of transepithelial resistance of Detroit 562 cell monolayers and fluorescence microscopy, leading to the assumption that C. diphtheriae strains might use epithelial cells as an environmental niche supplying protection against antibodies and macrophages.
Conclusions: The results obtained suggest that it is necessary to investigate various isolates on a molecular level to understand and to predict the colonization process of different C. diphtheriae strains.
Figures






Similar articles
-
Contour and persistence length of Corynebacterium diphtheriae pili by atomic force microscopy.Eur Biophys J. 2012 Jun;41(6):561-70. doi: 10.1007/s00249-012-0818-4. Epub 2012 May 16. Eur Biophys J. 2012. PMID: 22588485
-
Corynebacterium diphtheriae invasion-associated protein (DIP1281) is involved in cell surface organization, adhesion and internalization in epithelial cells.BMC Microbiol. 2010 Jan 5;10:2. doi: 10.1186/1471-2180-10-2. BMC Microbiol. 2010. PMID: 20051108 Free PMC article.
-
Induction of the NFκ-B signal transduction pathway in response to Corynebacterium diphtheriae infection.Microbiology (Reading). 2013 Jan;159(Pt 1):126-135. doi: 10.1099/mic.0.061879-0. Epub 2012 Nov 1. Microbiology (Reading). 2013. PMID: 23125120
-
[Adhesion of corynebacterium diphtheriae: the role of surface structures and formation mechanism].Zh Mikrobiol Epidemiol Immunobiol. 2014 Jul-Aug;(4):109-17. Zh Mikrobiol Epidemiol Immunobiol. 2014. PMID: 25286540 Review. Russian.
-
Adhesion by pathogenic corynebacteria.Adv Exp Med Biol. 2011;715:91-103. doi: 10.1007/978-94-007-0940-9_6. Adv Exp Med Biol. 2011. PMID: 21557059 Review.
Cited by
-
Cell envelope of corynebacteria: structure and influence on pathogenicity.ISRN Microbiol. 2013 Jan 21;2013:935736. doi: 10.1155/2013/935736. Print 2013. ISRN Microbiol. 2013. PMID: 23724339 Free PMC article.
-
The C-terminal coiled-coil domain of Corynebacterium diphtheriae DIP0733 is crucial for interaction with epithelial cells and pathogenicity in invertebrate animal model systems.BMC Microbiol. 2018 Sep 4;18(1):106. doi: 10.1186/s12866-018-1247-z. BMC Microbiol. 2018. PMID: 30180805 Free PMC article.
-
Contour and persistence length of Corynebacterium diphtheriae pili by atomic force microscopy.Eur Biophys J. 2012 Jun;41(6):561-70. doi: 10.1007/s00249-012-0818-4. Epub 2012 May 16. Eur Biophys J. 2012. PMID: 22588485
-
A multiomic approach to defining the essential genome of the globally important pathogen Corynebacterium diphtheriae.PLoS Genet. 2023 Apr 26;19(4):e1010737. doi: 10.1371/journal.pgen.1010737. eCollection 2023 Apr. PLoS Genet. 2023. PMID: 37099600 Free PMC article.
-
Genomic epidemiology of nontoxigenic Corynebacterium diphtheriae from King County, Washington State, USA between July 2018 and May 2019.Microb Genom. 2020 Dec;6(12):mgen000467. doi: 10.1099/mgen.0.000467. Epub 2020 Dec 4. Microb Genom. 2020. PMID: 33275088 Free PMC article.
References
-
- von Hunolstein C, Alfarone G, Scopetti F, Pataracchia M, La Valle R, Franchi F, Pacciani L, Manera A, Giammanco A, Farinelli S, Engler K, De Zoysa A, Efstratiou A. Molecular epidemiology and characteristics of Corynebacterium diphtheriae and Corynebacterium ulcerans strains isolated in Italy during the 1990s. J Med Microbiol. 2003;52:181–188. doi: 10.1099/jmm.0.04864-0. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

