A high-throughput cloning system for reverse genetics in Trypanosoma cruzi
- PMID: 20942965
- PMCID: PMC3020659
- DOI: 10.1186/1471-2180-10-259
A high-throughput cloning system for reverse genetics in Trypanosoma cruzi
Abstract
Background: The three trypanosomatids pathogenic to men, Trypanosoma cruzi, Trypanosoma brucei and Leishmania major, are etiological agents of Chagas disease, African sleeping sickness and cutaneous leishmaniasis, respectively. The complete sequencing of these trypanosomatid genomes represented a breakthrough in the understanding of these organisms. Genome sequencing is a step towards solving the parasite biology puzzle, as there are a high percentage of genes encoding proteins without functional annotation. Also, technical limitations in protein expression in heterologous systems reinforce the evident need for the development of a high-throughput reverse genetics platform. Ideally, such platform would lead to efficient cloning and compatibility with various approaches. Thus, we aimed to construct a highly efficient cloning platform compatible with plasmid vectors that are suitable for various approaches.
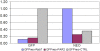
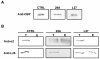
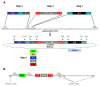
Results: We constructed a platform with a flexible structure allowing the exchange of various elements, such as promoters, fusion tags, intergenic regions or resistance markers. This platform is based on Gateway® technology, to ensure a fast and efficient cloning system. We obtained plasmid vectors carrying genes for fluorescent proteins (green, cyan or yellow), and sequences for the c-myc epitope, and tandem affinity purification or polyhistidine tags. The vectors were verified by successful subcellular localization of two previously characterized proteins (TcRab7 and PAR 2) and a putative centrin. For the tandem affinity purification tag, the purification of two protein complexes (ribosome and proteasome) was performed.
Conclusions: We constructed plasmids with an efficient cloning system and suitable for use across various applications, such as protein localization and co-localization, protein partner identification and protein expression. This platform also allows vector customization, as the vectors were constructed to enable easy exchange of its elements. The development of this high-throughput platform is a step closer towards large-scale trypanosome applications and initiatives.
Figures



References
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources

