The differential activity of interferon-α subtypes is consistent among distinct target genes and cell types
- PMID: 20943413
- PMCID: PMC3020287
- DOI: 10.1016/j.cyto.2010.09.006
The differential activity of interferon-α subtypes is consistent among distinct target genes and cell types
Abstract
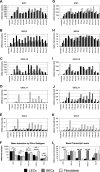
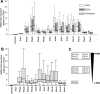
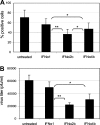
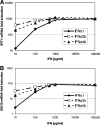
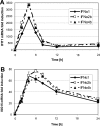
IFN-α proteins have been described to originate from 14 individual genes and allelic variants. However, the exceptional diversity of IFN-α and its functional impact are still poorly understood. To characterize the biological activity of IFN-α subtypes in relation to the cellular background, we investigated the effect of IFN-α treatment in primary fibroblasts and endothelial cells of vascular or lymphatic origin. The cellular response was evaluated for 13 distinct IFN-α proteins with respect to transcript regulation of the IFN-stimulated genes (ISGs) IFIT1, ISG15, CXCL10, CXCL11 and CCL8. The IFN-α proteins displayed a remarkably consistent potency in gene induction irrespective of target gene and cellular background which led to the classification of IFN-α subtypes with low (IFN-α1), intermediate (IFN-α2a, -4a, -4b, -5, -16, -21) and high (IFN-α2b, -6, -7, -8, -10, -14) activity. The differential potency of IFN-α classes was confirmed at the ISG protein level and the functional protection of cells against influenza virus infection. Differences in IFN activity were only observed at subsaturating levels of IFN-α proteins and did not affect the time course of ISG regulation. Cell-type specific responses were apparent for distinct target genes independent of IFN-α subtype and were based on different levels of basal versus inducible gene expression. While fibroblasts presented with a high constitutive level of IFIT1, the expression in endothelial cells was strongly induced by IFN-α. In contrast, CXCL10 and CXCL11 transcript levels were generally higher in endothelial cells despite a pronounced induction by IFN-α in fibroblasts. In summary, the divergent potency of IFN-α proteins and the cell-type specific regulation of individual IFN target genes may allow for the fine tuning of cellular responses to pathogen defense.
Copyright © 2010 Elsevier Ltd. All rights reserved.
Figures

Similar articles
-
Differential responses to IFN-alpha subtypes in human T cells and dendritic cells.J Immunol. 2003 Nov 15;171(10):5255-63. doi: 10.4049/jimmunol.171.10.5255. J Immunol. 2003. PMID: 14607926
-
Interferon-gamma potentiates the antiviral activity and the expression of interferon-stimulated genes induced by interferon-alpha in U937 cells.J Interferon Res. 1992 Apr;12(2):87-94. doi: 10.1089/jir.1992.12.87. J Interferon Res. 1992. PMID: 1315834
-
Antiviral potency analysis and functional comparison of consensus interferon, interferon-alpha2a and pegylated interferon-alpha2b against hepatitis C virus infection.Antivir Ther. 2008;13(7):851-62. Antivir Ther. 2008. PMID: 19043919 Free PMC article.
-
Dose-Dependent Differences in HIV Inhibition by Different Interferon Alpha Subtypes While Having Overall Similar Biologic Effects.mSphere. 2019 Feb 13;4(1):e00637-18. doi: 10.1128/mSphere.00637-18. mSphere. 2019. PMID: 30760614 Free PMC article.
-
Interferon-induced gene expression is a stronger predictor of treatment response than IL28B genotype in patients with hepatitis C.Gastroenterology. 2011 Mar;140(3):1021-31. doi: 10.1053/j.gastro.2010.11.039. Epub 2010 Nov 25. Gastroenterology. 2011. PMID: 21111740
Cited by
-
Dynamic YAP expression in the non-parenchymal liver cell compartment controls heterologous cell communication.Cell Mol Life Sci. 2024 Mar 4;81(1):115. doi: 10.1007/s00018-024-05126-1. Cell Mol Life Sci. 2024. PMID: 38436764 Free PMC article.
-
Type I interferon and mitochondrial dysfunction are associated with dysregulated cytotoxic CD8+ T cell responses in juvenile systemic lupus erythematosus.Clin Exp Immunol. 2025 Jan 21;219(1):uxae127. doi: 10.1093/cei/uxae127. Clin Exp Immunol. 2025. PMID: 39719886 Free PMC article.
-
CD8(+) lymphocytes suppress human immunodeficiency virus 1 replication by secreting type I interferons.J Interferon Cytokine Res. 2013 Nov;33(11):632-45. doi: 10.1089/jir.2012.0067. Epub 2013 Feb 12. J Interferon Cytokine Res. 2013. PMID: 23402527 Free PMC article.
-
Sex-dependent differences in type I IFN-induced natural killer cell activation.Front Immunol. 2023 Dec 15;14:1277967. doi: 10.3389/fimmu.2023.1277967. eCollection 2023. Front Immunol. 2023. PMID: 38162640 Free PMC article.
-
Plasmacytoid dendritic cells respond to Epstein-Barr virus infection with a distinct type I interferon subtype profile.Blood Adv. 2019 Apr 9;3(7):1129-1144. doi: 10.1182/bloodadvances.2018025536. Blood Adv. 2019. PMID: 30952679 Free PMC article.
References
-
- Isaacs A., Lindenmann J. Virus interference. I. The interferon. Proc R Soc Lond B Biol Sci. 1957;147:258–267. - PubMed
-
- Pestka S., Krause C.D., Walter M.R. Interferons, interferon-like cytokines, and their receptors. Immunol Rev. 2004;202:8–32. - PubMed
-
- Diaz M.O., Pomykala H.M., Bohlander S.K., Maltepe E., Malik K., Brownstein B. Structure of the human type-I interferon gene cluster determined from a YAC clone contig. Genomics. 1994;22:540–552. - PubMed
-
- Chen J., Baig E., Fish E.N. Diversity and relatedness among the type I interferons. J Interferon Cytokine Res. 2004;24:687–698. - PubMed
-
- Platanias L.C. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat Rev Immunol. 2005;5:375–386. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

