Activity and cellular functions of the deubiquitinating enzyme and polyglutamine disease protein ataxin-3 are regulated by ubiquitination at lysine 117
- PMID: 20943656
- PMCID: PMC2998082
- DOI: 10.1074/jbc.M110.181610
Activity and cellular functions of the deubiquitinating enzyme and polyglutamine disease protein ataxin-3 are regulated by ubiquitination at lysine 117
Abstract
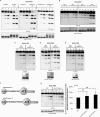
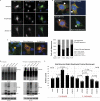
Deubiquitinating enzymes (DUbs) play important roles in many ubiquitin-dependent pathways, yet how DUbs themselves are regulated is not well understood. Here, we provide insight into the mechanism by which ubiquitination directly enhances the activity of ataxin-3, a DUb implicated in protein quality control and the disease protein in the polyglutamine neurodegenerative disorder, Spinocerebellar Ataxia Type 3. We identify Lys-117, which resides near the catalytic triad, as the primary site of ubiquitination in wild type and pathogenic ataxin-3. Further studies indicate that ubiquitin-dependent activation of ataxin-3 at Lys-117 is important for its ability to reduce high molecular weight ubiquitinated species in cells. Ubiquitination at Lys-117 also facilitates the ability of ataxin-3 to induce aggresome formation in cells. Finally, structure-function studies support a model of activation whereby ubiquitination at Lys-117 enhances ataxin-3 activity independent of the known ubiquitin-binding sites in ataxin-3, most likely through a direct conformational change in or near the catalytic domain.
Figures




Similar articles
-
Ubiquitination directly enhances activity of the deubiquitinating enzyme ataxin-3.EMBO J. 2009 Feb 18;28(4):372-82. doi: 10.1038/emboj.2008.289. Epub 2009 Jan 15. EMBO J. 2009. PMID: 19153604 Free PMC article.
-
Ubiquitination regulates the neuroprotective function of the deubiquitinase ataxin-3 in vivo.J Biol Chem. 2013 Nov 29;288(48):34460-9. doi: 10.1074/jbc.M113.513903. Epub 2013 Oct 8. J Biol Chem. 2013. PMID: 24106274 Free PMC article.
-
Understanding the role of the Josephin domain in the PolyUb binding and cleavage properties of ataxin-3.PLoS One. 2010 Aug 26;5(8):e12430. doi: 10.1371/journal.pone.0012430. PLoS One. 2010. PMID: 20865150 Free PMC article.
-
Polyglutamine diseases: the special case of ataxin-3 and Machado-Joseph disease.Prog Neurobiol. 2011 Sep 15;95(1):26-48. doi: 10.1016/j.pneurobio.2011.06.007. Epub 2011 Jun 28. Prog Neurobiol. 2011. PMID: 21740957 Review.
-
The deubiquitinase function of ataxin-3 and its role in the pathogenesis of Machado-Joseph disease and other diseases.Biochem J. 2024 Mar 20;481(6):461-480. doi: 10.1042/BCJ20240017. Biochem J. 2024. PMID: 38497605 Free PMC article. Review.
Cited by
-
Ubiquitin receptors and protein quality control.J Mol Cell Cardiol. 2013 Feb;55:73-84. doi: 10.1016/j.yjmcc.2012.09.012. Epub 2012 Oct 6. J Mol Cell Cardiol. 2013. PMID: 23046644 Free PMC article. Review.
-
Ubiquitin-specific protease 25 functions in Endoplasmic Reticulum-associated degradation.PLoS One. 2012;7(5):e36542. doi: 10.1371/journal.pone.0036542. Epub 2012 May 9. PLoS One. 2012. PMID: 22590560 Free PMC article.
-
Role of the ubiquitin-proteasome system in nervous system function and disease: using C. elegans as a dissecting tool.Cell Mol Life Sci. 2012 Aug;69(16):2691-715. doi: 10.1007/s00018-012-0946-0. Epub 2012 Mar 3. Cell Mol Life Sci. 2012. PMID: 22382927 Free PMC article. Review.
-
Ataxin-3 protein and RNA toxicity in spinocerebellar ataxia type 3: current insights and emerging therapeutic strategies.Mol Neurobiol. 2014 Jun;49(3):1513-31. doi: 10.1007/s12035-013-8596-2. Epub 2013 Nov 29. Mol Neurobiol. 2014. PMID: 24293103 Free PMC article. Review.
-
Differential toxicity of ataxin-3 isoforms in Drosophila models of Spinocerebellar Ataxia Type 3.Neurobiol Dis. 2019 Dec;132:104535. doi: 10.1016/j.nbd.2019.104535. Epub 2019 Jul 13. Neurobiol Dis. 2019. PMID: 31310802 Free PMC article.
References
-
- Amerik A. Y., Hochstrasser M. (2004) Biochim. Biophys Acta 1695, 189–207 - PubMed
-
- Komander D., Clague M. J., Urbé S. (2009) Nat. Rev. Mol. Cell. Biol. 10, 550–563 - PubMed
-
- Nijman S. M., Luna-Vargas M. P., Velds A., Brummelkamp T. R., Dirac A. M., Sixma T. K., Bernards R. (2005) Cell 123, 773–786 - PubMed
-
- Fischer J. A. (2003) Int. Rev. Cytol. 229, 43–72 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

