The dyslexia-associated KIAA0319 protein undergoes proteolytic processing with {gamma}-secretase-independent intramembrane cleavage
- PMID: 20943657
- PMCID: PMC3000997
- DOI: 10.1074/jbc.M110.145961
The dyslexia-associated KIAA0319 protein undergoes proteolytic processing with {gamma}-secretase-independent intramembrane cleavage
Abstract
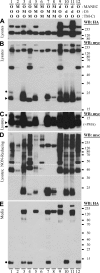
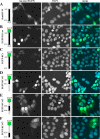
The KIAA0319 gene has been associated with reading disability in several studies. It encodes a plasma membrane protein with a large, highly glycosylated, extracellular domain. This protein is proposed to function in adhesion and attachment and thought to play an important role during neuronal migration in the developing brain. We have previously proposed that endocytosis of this protein could constitute an important mechanism to regulate its function. Here we show that KIAA0319 undergoes ectodomain shedding and intramembrane cleavage. At least five different cleavage events occur, four in the extracellular domain and one within the transmembrane domain. The ectodomain shedding processing cleaves the extracellular domain, generating several small fragments, including the N-terminal region with the Cys-rich MANEC domain. It is possible that these fragments are released to the extracellular medium and trigger cellular responses. The intramembrane cleavage releases the intracellular domain from its membrane attachment. Our results suggest that this cleavage event is not carried out by γ-secretase, the enzyme complex involved in similar processing in many other type I proteins. The soluble cytoplasmic domain of KIAA0319 is able to translocate to the nucleus, accumulating in nucleoli after overexpression. This fragment has an unknown role, although it could be involved in regulation of gene expression. The absence of DNA-interacting motifs indicates that such a function would most probably be mediated through interaction with other proteins, not by direct DNA binding. These results suggest that KIAA0319 not only has a direct role in neuronal migration but may also have additional signaling functions.
Figures






Similar articles
-
Normal radial migration and lamination are maintained in dyslexia-susceptibility candidate gene homolog Kiaa0319 knockout mice.Brain Struct Funct. 2017 Apr;222(3):1367-1384. doi: 10.1007/s00429-016-1282-1. Epub 2016 Aug 10. Brain Struct Funct. 2017. PMID: 27510895 Free PMC article.
-
The dyslexia-associated protein KIAA0319 interacts with adaptor protein 2 and follows the classical clathrin-mediated endocytosis pathway.Am J Physiol Cell Physiol. 2009 Jul;297(1):C160-8. doi: 10.1152/ajpcell.00630.2008. Epub 2009 May 6. Am J Physiol Cell Physiol. 2009. PMID: 19419997 Free PMC article.
-
The dyslexia-associated gene KIAA0319 encodes highly N- and O-glycosylated plasma membrane and secreted isoforms.Hum Mol Genet. 2008 Mar 15;17(6):859-71. doi: 10.1093/hmg/ddm358. Epub 2007 Dec 6. Hum Mol Genet. 2008. PMID: 18063668
-
Gamma-secretase-dependent signaling of receptor tyrosine kinases.Oncogene. 2019 Jan;38(2):151-163. doi: 10.1038/s41388-018-0465-z. Epub 2018 Aug 30. Oncogene. 2019. PMID: 30166589 Free PMC article. Review.
-
AICD nuclear signaling and its possible contribution to Alzheimer's disease.Curr Alzheimer Res. 2012 Feb;9(2):200-16. doi: 10.2174/156720512799361673. Curr Alzheimer Res. 2012. PMID: 21605035 Review.
Cited by
-
KIAA0319 gene polymorphisms are associated with developmental dyslexia in Chinese Uyghur children.J Hum Genet. 2016 Aug;61(8):745-52. doi: 10.1038/jhg.2016.40. Epub 2016 Apr 21. J Hum Genet. 2016. PMID: 27098879 Free PMC article.
-
[Research advances in susceptible genes for developmental dyslexia in children].Zhongguo Dang Dai Er Ke Za Zhi. 2016 Dec;18(12):1308-1312. doi: 10.7499/j.issn.1008-8830.2016.12.022. Zhongguo Dang Dai Er Ke Za Zhi. 2016. PMID: 27974128 Free PMC article. Review. Chinese.
-
The genetics of reading disabilities: from phenotypes to candidate genes.Front Psychol. 2013 Jan 7;3:601. doi: 10.3389/fpsyg.2012.00601. eCollection 2012. Front Psychol. 2013. PMID: 23308072 Free PMC article.
-
KIAA0319 influences cilia length, cell migration and mechanical cell-substrate interaction.Sci Rep. 2022 Jan 14;12(1):722. doi: 10.1038/s41598-021-04539-3. Sci Rep. 2022. PMID: 35031635 Free PMC article.
-
Knockdown of the dyslexia-associated gene Kiaa0319 impairs temporal responses to speech stimuli in rat primary auditory cortex.Cereb Cortex. 2014 Jul;24(7):1753-66. doi: 10.1093/cercor/bht028. Epub 2013 Feb 8. Cereb Cortex. 2014. PMID: 23395846 Free PMC article.
References
-
- Paracchini S., Scerri T., Monaco A. P. (2007) Annu. Rev. Genomics Hum. Genet. 8, 57–79 - PubMed
-
- Fisher S. E., Francks C. (2006) Trends Cogn. Sci. 10, 250–257 - PubMed
-
- McGrath L. M., Smith S. D., Pennington B. F. (2006) Trends Mol. Med. 12, 333–341 - PubMed
-
- Galaburda A. M., LoTurco J., Ramus F., Fitch R. H., Rosen G. D. (2006) Nat. Neurosci. 9, 1213–1217 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

