Crystal structure of cardiac-specific histone methyltransferase SmyD1 reveals unusual active site architecture
- PMID: 20943667
- PMCID: PMC3003362
- DOI: 10.1074/jbc.M110.168187
Crystal structure of cardiac-specific histone methyltransferase SmyD1 reveals unusual active site architecture
Abstract
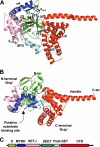
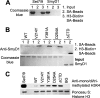
SmyD1 is a cardiac- and muscle-specific histone methyltransferase that methylates histone H3 at lysine 4 and regulates gene transcription in early heart development. The unique domain structure characterized by a "split" SET domain, a conserved MYND zinc finger, and a novel C-terminal domain (CTD) distinguishes SmyD1 from other SET domain containing methyltransferases. Here we report the crystal structure of full-length SmyD1 in complex with the cofactor analog sinefungin at 2.3 Å. The structure reveals that SmyD1 folds into a wrench-shaped structure with two thick "grips" separated by a large, deep concave opening. Importantly, our structural and functional analysis suggests that SmyD1 appears to be regulated by an autoinhibition mechanism, and that unusually spacious target lysine-access channel and the presence of the CTD domain both negatively contribute to the regulation of this cardiovascularly relevant methyltransferase. Furthermore, our structure also provides a structural basis for the interaction between SmyD1 and cardiac transcription factor skNAC, and suggests that the MYND domain may primarily serve as a protein interaction module and cooperate SmyD1 with skNAC to regulate cardiomyocyte growth and maturation. Overall, our data provide novel insights into the mechanism of SmyD1 regulation, which would be helpful in further understanding the role of this protein in heart development and cardiovascular diseases.
Figures



Similar articles
-
Lysine Methyltransferase SMYD1 Regulates Myogenesis via skNAC Methylation.Cells. 2023 Jun 22;12(13):1695. doi: 10.3390/cells12131695. Cells. 2023. PMID: 37443729 Free PMC article.
-
Histone methyltransferase Smyd1 regulates mitochondrial energetics in the heart.Proc Natl Acad Sci U S A. 2018 Aug 14;115(33):E7871-E7880. doi: 10.1073/pnas.1800680115. Epub 2018 Jul 30. Proc Natl Acad Sci U S A. 2018. PMID: 30061404 Free PMC article.
-
skNAC, a Smyd1-interacting transcription factor, is involved in cardiac development and skeletal muscle growth and regeneration.Proc Natl Acad Sci U S A. 2010 Nov 30;107(48):20750-5. doi: 10.1073/pnas.1013493107. Epub 2010 Nov 11. Proc Natl Acad Sci U S A. 2010. PMID: 21071677 Free PMC article.
-
Structure and function of SET and MYND domain-containing proteins.Int J Mol Sci. 2015 Jan 8;16(1):1406-28. doi: 10.3390/ijms16011406. Int J Mol Sci. 2015. PMID: 25580534 Free PMC article. Review.
-
SMYD proteins: key regulators in skeletal and cardiac muscle development and function.Anat Rec (Hoboken). 2014 Sep;297(9):1650-62. doi: 10.1002/ar.22972. Anat Rec (Hoboken). 2014. PMID: 25125178 Review.
Cited by
-
Structural and functional analysis of the DEAF-1 and BS69 MYND domains.PLoS One. 2013;8(1):e54715. doi: 10.1371/journal.pone.0054715. Epub 2013 Jan 25. PLoS One. 2013. PMID: 23372760 Free PMC article.
-
Protein crystallization: Eluding the bottleneck of X-ray crystallography.AIMS Biophys. 2017;4(4):557-575. doi: 10.3934/biophy.2017.4.557. Epub 2017 Sep 26. AIMS Biophys. 2017. PMID: 29051919 Free PMC article.
-
Structure of the SMYD2-PARP1 Complex Reveals Both Productive and Allosteric Modes of Peptide Binding.bioRxiv [Preprint]. 2024 Dec 4:2024.12.03.626679. doi: 10.1101/2024.12.03.626679. bioRxiv. 2024. PMID: 39677743 Free PMC article. Preprint.
-
Master redox regulator Trx1 upregulates SMYD1 & modulates lysine methylation.Biochim Biophys Acta. 2015 Dec;1854(12):1816-1822. doi: 10.1016/j.bbapap.2015.09.006. Epub 2015 Sep 26. Biochim Biophys Acta. 2015. PMID: 26410624 Free PMC article.
-
Lysine Methyltransferase SMYD1 Regulates Myogenesis via skNAC Methylation.Cells. 2023 Jun 22;12(13):1695. doi: 10.3390/cells12131695. Cells. 2023. PMID: 37443729 Free PMC article.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

