Membrane anchoring subunits specify selective regulation of RGS9·Gbeta5 GAP complex in photoreceptor neurons
- PMID: 20943919
- PMCID: PMC2975674
- DOI: 10.1523/JNEUROSCI.1191-10.2010
Membrane anchoring subunits specify selective regulation of RGS9·Gbeta5 GAP complex in photoreceptor neurons
Abstract
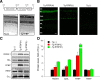
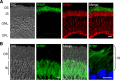
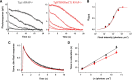
The RGS9·Gβ5 complex is the key regulator of neuronal G-protein signaling and shows remarkable selectivity of subunit composition. In retinal photoreceptors, RGS9·Gβ5 is bound to the membrane anchor R9AP and the complex regulates visual signaling. In the basal ganglia neurons, RGS9·Gβ5 is instead associated with a homologous protein, R7BP, and regulates reward circuit. Switching this selective subunit composition of the complex in rod photoreceptors allowed us to study the molecular underpinning of signaling specificity in diverse G-protein pathways. We have found that both membrane anchoring subunits play a conserved role in regulating protein levels of RGS9·Gβ5 and enhancing the ability of RGS·Gβ5 complexes to stimulate GTPase activity of G proteins. However, notable differences exist in the subcellular targeting of alternatively configured complexes. Unlike R9AP, which relies on passive targeting mechanisms for the delivery to the outer segments of the photoreceptors, R7BP is excluded from this location and is instead specifically targeted to the plasma membrane. R7BP-containing complexes could be rerouted to the outer segments, where they are capable of regulating the phototransduction cascade by the active targeting signals derived from rhodopsin. These findings illustrate the diversity of the G-protein signaling regulation by RGS·Gβ5 complexes achieved by differential recruitment of the membrane anchors.
Figures




Similar articles
-
Targeting of RGS7/Gbeta5 to the dendritic tips of ON-bipolar cells is independent of its association with membrane anchor R7BP.J Neurosci. 2008 Oct 8;28(41):10443-9. doi: 10.1523/JNEUROSCI.3282-08.2008. J Neurosci. 2008. PMID: 18842904 Free PMC article.
-
Membrane attachment is key to protecting transducin GTPase-activating complex from intracellular proteolysis in photoreceptors.J Neurosci. 2011 Oct 12;31(41):14660-8. doi: 10.1523/JNEUROSCI.3516-11.2011. J Neurosci. 2011. PMID: 21994382 Free PMC article.
-
Absence of the RGS9.Gbeta5 GTPase-activating complex in photoreceptors of the R9AP knockout mouse.J Biol Chem. 2004 Jan 16;279(3):1581-4. doi: 10.1074/jbc.C300456200. Epub 2003 Nov 18. J Biol Chem. 2004. PMID: 14625292
-
R9AP and R7BP: traffic cops for the RGS7 family in phototransduction and neuronal GPCR signaling.Trends Pharmacol Sci. 2009 Jan;30(1):17-24. doi: 10.1016/j.tips.2008.10.002. Epub 2008 Nov 29. Trends Pharmacol Sci. 2009. PMID: 19042037 Free PMC article. Review.
-
Structure, function, and localization of Gβ5-RGS complexes.Prog Mol Biol Transl Sci. 2009;86:157-203. doi: 10.1016/S1877-1173(09)86006-7. Epub 2009 Oct 7. Prog Mol Biol Transl Sci. 2009. PMID: 20374716 Free PMC article. Review.
Cited by
-
Effect of Rhodopsin Phosphorylation on Dark Adaptation in Mouse Rods.J Neurosci. 2016 Jun 29;36(26):6973-87. doi: 10.1523/JNEUROSCI.3544-15.2016. J Neurosci. 2016. PMID: 27358455 Free PMC article.
-
G protein signaling in the retina and beyond: the Cogan lecture.Invest Ophthalmol Vis Sci. 2014 Dec 15;55(12):8201-7. doi: 10.1167/iovs.14-15928. Invest Ophthalmol Vis Sci. 2014. PMID: 25511392 Free PMC article. No abstract available.
-
Type 5 G protein beta subunit (Gbeta5) controls the interaction of regulator of G protein signaling 9 (RGS9) with membrane anchors.J Biol Chem. 2011 Jun 17;286(24):21806-13. doi: 10.1074/jbc.M111.241513. Epub 2011 Apr 21. J Biol Chem. 2011. PMID: 21511947 Free PMC article.
-
Differential function of Gγ13 in rod bipolar and ON cone bipolar cells.J Physiol. 2015 Apr 1;593(7):1531-50. doi: 10.1113/jphysiol.2014.281196. Epub 2015 Jan 2. J Physiol. 2015. PMID: 25416620 Free PMC article.
-
Photoreceptors in whirler mice show defective transducin translocation and are susceptible to short-term light/dark changes-induced degeneration.Exp Eye Res. 2014 Jan;118:145-53. doi: 10.1016/j.exer.2013.10.021. Epub 2013 Nov 7. Exp Eye Res. 2014. PMID: 24211856 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
