Slow degradation and aggregation in vitro of mutant GABAA receptor gamma2(Q351X) subunits associated with epilepsy
- PMID: 20943930
- PMCID: PMC2976503
- DOI: 10.1523/JNEUROSCI.2320-10.2010
Slow degradation and aggregation in vitro of mutant GABAA receptor gamma2(Q351X) subunits associated with epilepsy
Abstract
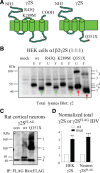
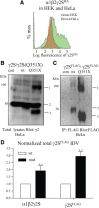
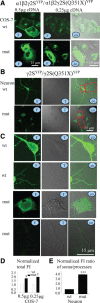
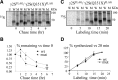
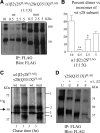
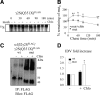
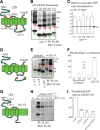
The GABA(A) receptor γ2 subunit nonsense mutation Q351X has been associated with the genetic epilepsy syndrome generalized epilepsy with febrile seizures plus, which includes a spectrum of seizures types from febrile seizures to Dravet syndrome. Although most genetic epilepsy syndromes are mild and remit with age, Dravet syndrome has a more severe clinical course with refractory seizures associated with developmental delay and cognitive impairment. The basis for the broad spectrum of seizure phenotypes is uncertain. We demonstrated previously that the GABA(A) receptor γ2 subunit gene Q351X mutation suppressed biogenesis of wild-type partnering α1 and β2 subunits in addition to its loss of function. Here we show that γ2S(Q351X) subunits have an additional impairment of biogenesis. Mutant γ2(Q351X) subunits were degraded more slowly than wild-type γ2 subunits and formed SDS-resistant, high-molecular-mass complexes or aggregates in multiple cell types, including neurons. The half-life of γ2S(Q351X) subunits was ∼4 h, whereas that of γ2S subunits was ∼2 h. Mutant subunits formed complexes rapidly after synthesis onset. Using multiple truncated subunits, we demonstrated that aggregate formation was a general phenomenon for truncated γ2S subunits and that their Cys-loop cysteines were involved in aggregate formation. Protein aggregation is a hallmark of neurodegenerative diseases, but the effects of the mutant γ2S(Q351X) subunit aggregates on neuronal function and survival are unclear. Additional validation of the mutant subunit aggregation in vivo and determination of the involved signaling pathways will help reveal the pathological effects of these mutant subunit aggregates in the pathogenesis of genetic epilepsy syndromes.
Figures

Similar articles
-
The GABRG2 mutation, Q351X, associated with generalized epilepsy with febrile seizures plus, has both loss of function and dominant-negative suppression.J Neurosci. 2009 Mar 4;29(9):2845-56. doi: 10.1523/JNEUROSCI.4772-08.2009. J Neurosci. 2009. PMID: 19261880 Free PMC article.
-
Overexpressing wild-type γ2 subunits rescued the seizure phenotype in Gabrg2+/Q390X Dravet syndrome mice.Epilepsia. 2017 Aug;58(8):1451-1461. doi: 10.1111/epi.13810. Epub 2017 Jun 6. Epilepsia. 2017. PMID: 28586508 Free PMC article.
-
Why does fever trigger febrile seizures? GABAA receptor gamma2 subunit mutations associated with idiopathic generalized epilepsies have temperature-dependent trafficking deficiencies.J Neurosci. 2006 Mar 1;26(9):2590-7. doi: 10.1523/JNEUROSCI.4243-05.2006. J Neurosci. 2006. PMID: 16510738 Free PMC article.
-
Molecular Pathogenic Basis for GABRG2 Mutations Associated With a Spectrum of Epilepsy Syndromes, From Generalized Absence Epilepsy to Dravet Syndrome.JAMA Neurol. 2016 Aug 1;73(8):1009-16. doi: 10.1001/jamaneurol.2016.0449. JAMA Neurol. 2016. PMID: 27367160 Free PMC article. Review.
-
GABA(A) receptor epilepsy mutations.Biochem Pharmacol. 2004 Oct 15;68(8):1497-506. doi: 10.1016/j.bcp.2004.07.029. Biochem Pharmacol. 2004. PMID: 15451392 Review.
Cited by
-
Variable Expression of GABAA Receptor Subunit Gamma 2 Mutation in a Nuclear Family Displaying Developmental and Encephalopathic Phenotype.Int J Mol Sci. 2022 Aug 26;23(17):9683. doi: 10.3390/ijms23179683. Int J Mol Sci. 2022. PMID: 36077081 Free PMC article. Review.
-
De novo GABRG2 mutations associated with epileptic encephalopathies.Brain. 2017 Jan;140(1):49-67. doi: 10.1093/brain/aww272. Epub 2016 Nov 17. Brain. 2017. PMID: 27864268 Free PMC article.
-
Differential protein structural disturbances and suppression of assembly partners produced by nonsense GABRG2 epilepsy mutations: implications for disease phenotypic heterogeneity.Sci Rep. 2016 Oct 20;6:35294. doi: 10.1038/srep35294. Sci Rep. 2016. PMID: 27762395 Free PMC article.
-
4-Phenylbutyrate restored γ-aminobutyric acid uptake and reduced seizures in SLC6A1 patient variant-bearing cell and mouse models.Brain Commun. 2022 Jun 6;4(3):fcac144. doi: 10.1093/braincomms/fcac144. eCollection 2022. Brain Commun. 2022. PMID: 35911425 Free PMC article.
-
Altered GABA signaling in early life epilepsies.Neural Plast. 2011;2011:527605. doi: 10.1155/2011/527605. Epub 2011 Jul 31. Neural Plast. 2011. PMID: 21826277 Free PMC article. Review.
References
-
- Baulac S, Huberfeld G, Gourfinkel-An I, Mitropoulou G, Beranger A, Prud'homme JF, Baulac M, Brice A, Bruzzone R, LeGuern E. First genetic evidence of GABAA receptor dysfunction in epilepsy: a mutation in the gamma2-subunit gene. Nat Genet. 2001;28:46–48. - PubMed
-
- Bennett MC, Bishop JF, Leng Y, Chock PB, Chase TN, Mouradian MM. Degradation of alpha-synuclein by proteasome. J Biol Chem. 1999;274:33855–33858. - PubMed
-
- Berke SJ, Paulson HL. Protein aggregation and the ubiquitin proteasome pathway: gaining the UPPer hand on neurodegeneration. Curr Opin Genet Dev. 2003;13:253–261. - PubMed
-
- Berkovic SF, Harkin L, McMahon JM, Pelekanos JT, Zuberi SM, Wirrell EC, Gill DS, Iona X, Mulley JC, Scheffer IE. De-novo mutations of the sodium channel gene SCN1A in alleged vaccine encephalopathy: a retrospective study. Lancet Neurol. 2006;5:488–492. - PubMed
-
- Cohen-Gadol AA, Bradley CC, Williamson A, Kim JH, Westerveld M, Duckrow RB, Spencer DD. Normal magnetic resonance imaging and medial temporal lobe epilepsy: the clinical syndrome of paradoxical temporal lobe epilepsy. J Neurosurg. 2005;102:902–909. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
