Microautophagy of the nucleus coincides with a vacuolar diffusion barrier at nuclear-vacuolar junctions
- PMID: 20943953
- PMCID: PMC2993746
- DOI: 10.1091/mbc.E09-09-0782
Microautophagy of the nucleus coincides with a vacuolar diffusion barrier at nuclear-vacuolar junctions
Abstract
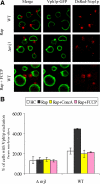
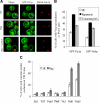
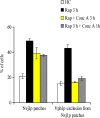
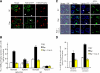
Nuclei bind yeast vacuoles via nucleus-vacuole (NV) junctions. Under nutrient restriction, NV junctions invaginate and release vesicles filled with nuclear material into vacuoles, resulting in piecemeal microautophagy of the nucleus (PMN). We show that the electrochemical gradient across the vacuolar membrane promotes invagination of NV junctions. Existing invaginations persist independently of the gradient, but final release of PMN vesicles requires again V-ATPase activity. We find that NV junctions form a diffusion barrier on the vacuolar membrane that excludes V-ATPase but is enriched in the VTC complex and accessible to other membrane-integral proteins. V-ATPase exclusion depends on the NV junction proteins Nvj1p,Vac8p, and the electrochemical gradient. It also depends on factors of lipid metabolism, such as the oxysterol binding protein Osh1p and the enoyl-CoA reductase Tsc13p, which are enriched in NV junctions, and on Lag1p and Fen1p. Our observations suggest that NV junctions form in two separable steps: Nvj1p and Vac8p suffice to establish contact between the two membranes. The electrochemical potential and lipid-modifying enzymes are needed to establish the vacuolar diffusion barrier, invaginate NV junctions, and form PMN vesicles.
Figures
References
-
- Ashman W. H., Barbuch R. J., Ulbright C. E., Jarrett H. W., Bard M. Cloning and disruption of the yeast C-8 sterol isomerase gene. Lipids. 1991;26:628–632. - PubMed
-
- Beh C. T., Rine J. A role for yeast oxysterol-binding protein homologs in endocytosis and in the maintenance of intracellular sterol-lipid distribution. J. Cell Sci. 2004;117:2983–2996. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

