Determinants of strain-specific differences in efficiency of reovirus entry
- PMID: 20943982
- PMCID: PMC3004336
- DOI: 10.1128/JVI.01385-10
Determinants of strain-specific differences in efficiency of reovirus entry
Abstract
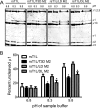
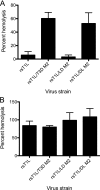
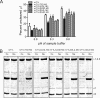
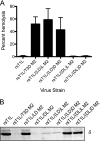
Cell entry of reovirus requires a series of ordered steps, which include conformational changes in outer capsid protein μ1 and its autocleavage. The μ1N fragment released as a consequence of these events interacts with host cell membranes and mediates their disruption, leading to delivery of the viral core into the cytoplasm. The prototype reovirus strains T1L and T3D exhibit differences in the efficiency of autocleavage, in the propensity to undergo conformational changes required for membrane penetration, and in the capacity for penetrating host cell membranes. To better understand how polymorphic differences in μ1 influence reovirus entry events, we generated recombinant viruses that express chimeric T1L-T3D μ1 proteins and characterized them for the capacity to efficiently complete each step required for membrane penetration. Our studies revealed two important functions for the central δ region of μ1. First, we found that μ1 autocleavage is regulated by the N-terminal portion of δ, which forms an α-helical pedestal structure. Second, we observed that the C-terminal portion of δ, which forms a jelly-roll β barrel structure, regulates membrane penetration by influencing the efficiency of ISVP* formation. Thus, our studies highlight the molecular basis for differences in the membrane penetration efficiency displayed by prototype reovirus strains and suggest that distinct portions of the reovirus δ domain influence different steps during entry.
Figures



Similar articles
-
Reovirus Core Proteins λ1 and σ2 Promote Stability of Disassembly Intermediates and Influence Early Replication Events.J Virol. 2020 Aug 17;94(17):e00491-20. doi: 10.1128/JVI.00491-20. Print 2020 Aug 17. J Virol. 2020. PMID: 32581098 Free PMC article.
-
Cleavage of the C-Terminal Fragment of Reovirus μ1 Is Required for Optimal Infectivity.J Virol. 2018 Feb 26;92(6):e01848-17. doi: 10.1128/JVI.01848-17. Print 2018 Mar 15. J Virol. 2018. PMID: 29298891 Free PMC article.
-
Lipids Cooperate with the Reovirus Membrane Penetration Peptide to Facilitate Particle Uncoating.J Biol Chem. 2016 Dec 23;291(52):26773-26785. doi: 10.1074/jbc.M116.747477. Epub 2016 Nov 15. J Biol Chem. 2016. PMID: 27875299 Free PMC article.
-
From touchdown to transcription: the reovirus cell entry pathway.Curr Top Microbiol Immunol. 2010;343:91-119. doi: 10.1007/82_2010_32. Curr Top Microbiol Immunol. 2010. PMID: 20397070 Free PMC article. Review.
-
Control of Capsid Transformations during Reovirus Entry.Viruses. 2021 Jan 21;13(2):153. doi: 10.3390/v13020153. Viruses. 2021. PMID: 33494426 Free PMC article. Review.
Cited by
-
Cell entry-associated conformational changes in reovirus particles are controlled by host protease activity.J Virol. 2012 Apr;86(7):3466-73. doi: 10.1128/JVI.06659-11. Epub 2012 Jan 25. J Virol. 2012. PMID: 22278245 Free PMC article.
-
Reovirus μ1 Protein Affects Infectivity by Altering Virus-Receptor Interactions.J Virol. 2016 Nov 14;90(23):10951-10962. doi: 10.1128/JVI.01843-16. Print 2016 Dec 1. J Virol. 2016. PMID: 27681135 Free PMC article.
-
Protein Mismatches Caused by Reassortment Influence Functions of the Reovirus Capsid.J Virol. 2018 Sep 26;92(20):e00858-18. doi: 10.1128/JVI.00858-18. Print 2018 Oct 15. J Virol. 2018. PMID: 30068646 Free PMC article.
-
Infectious Subviral Particle-induced Hemolysis Assay for Mammalian Orthoreovirus.Bio Protoc. 2018 Jan 20;8(2):e2701. doi: 10.21769/BioProtoc.2701. Bio Protoc. 2018. PMID: 29552594 Free PMC article.
-
Reovirus Core Proteins λ1 and σ2 Promote Stability of Disassembly Intermediates and Influence Early Replication Events.J Virol. 2020 Aug 17;94(17):e00491-20. doi: 10.1128/JVI.00491-20. Print 2020 Aug 17. J Virol. 2020. PMID: 32581098 Free PMC article.
References
-
- Banerjee, M., and J. E. Johnson. 2008. Activation, exposure and penetration of virally encoded, membrane-active polypeptides during non-enveloped virus entry. Curr. Protein Peptide Sci. 9:16-27. - PubMed
-
- Barton, E. S., J. L. Connolly, J. C. Forrest, J. D. Chappell, and T. S. Dermody. 2001. Utilization of sialic acid as a coreceptor enhances reovirus attachment by multistep adhesion strengthening. J. Biol. Chem. 276:2200-2211. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

