Structural basis of local, pH-dependent conformational changes in glycoprotein B from herpes simplex virus type 1
- PMID: 20943984
- PMCID: PMC3004323
- DOI: 10.1128/JVI.01750-10
Structural basis of local, pH-dependent conformational changes in glycoprotein B from herpes simplex virus type 1
Abstract
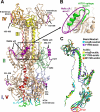
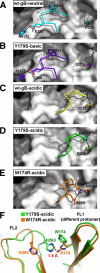
Herpesviruses enter cells by membrane fusion either at the plasma membrane or in endosomes, depending on the cell type. Glycoprotein B (gB) is a conserved component of the multiprotein herpesvirus fusion machinery and functions as a fusion protein, with two internal fusion loops, FL1 and FL2. We determined the crystal structures of the ectodomains of two FL1 mutants of herpes simplex virus type 1 (HSV-1) gB to clarify whether their fusion-null phenotypes were due to global or local effects of the mutations on the structure of the gB ectodomain. Each mutant has a single point mutation of a hydrophobic residue in FL1 that eliminates the hydrophobic side chain. We found that neither mutation affected the conformation of FL1, although one mutation slightly altered the conformation of FL2, and we conclude that the fusion-null phenotype is due to the absence of a hydrophobic side chain at the mutated position. Because the ectodomains of the wild-type and the mutant forms of gB crystallized at both low and neutral pH, we were able to determine the effect of pH on gB conformation at the atomic level. For viruses that enter cells by endocytosis, the low pH of the endosome effects major conformational changes in their fusion proteins, thereby promoting fusion of the viral envelope with the endosomal membrane. We show here that upon exposure of gB to low pH, FL2 undergoes a major relocation, probably driven by protonation of a key histidine residue. Relocation of FL2, as well as additional small conformational changes in the gB ectodomain, helps explain previously noted changes in its antigenic and biochemical properties. However, no global pH-dependent changes in gB structure were detected in either the wild-type or the mutant forms of gB. Thus, low pH causes local conformational changes in gB that are very different from the large-scale fusogenic conformational changes in other viral fusion proteins. We propose that these conformational changes, albeit modest, play an important functional role during endocytic entry of HSV.
Figures
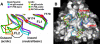


Similar articles
-
Mildly Acidic pH Triggers an Irreversible Conformational Change in the Fusion Domain of Herpes Simplex Virus 1 Glycoprotein B and Inactivation of Viral Entry.J Virol. 2017 Feb 14;91(5):e02123-16. doi: 10.1128/JVI.02123-16. Print 2017 Mar 1. J Virol. 2017. PMID: 28003487 Free PMC article.
-
Fusion-deficient insertion mutants of herpes simplex virus type 1 glycoprotein B adopt the trimeric postfusion conformation.J Virol. 2010 Feb;84(4):2001-12. doi: 10.1128/JVI.01791-09. Epub 2009 Nov 25. J Virol. 2010. PMID: 19939928 Free PMC article.
-
Acidic pH Mediates Changes in Antigenic and Oligomeric Conformation of Herpes Simplex Virus gB and Is a Determinant of Cell-Specific Entry.J Virol. 2018 Aug 16;92(17):e01034-18. doi: 10.1128/JVI.01034-18. Print 2018 Sep 1. J Virol. 2018. PMID: 29925660 Free PMC article.
-
Herpesvirus Entry into Host Cells Mediated by Endosomal Low pH.Traffic. 2016 Sep;17(9):965-75. doi: 10.1111/tra.12408. Epub 2016 May 24. Traffic. 2016. PMID: 27126894 Free PMC article. Review.
-
Two Sides to Every Story: Herpes Simplex Type-1 Viral Glycoproteins gB, gD, gH/gL, gK, and Cellular Receptors Function as Key Players in Membrane Fusion.Viruses. 2021 Sep 16;13(9):1849. doi: 10.3390/v13091849. Viruses. 2021. PMID: 34578430 Free PMC article. Review.
Cited by
-
The membrane-proximal region (MPR) of herpes simplex virus gB regulates association of the fusion loops with lipid membranes.mBio. 2012 Nov 20;3(6):e00429-12. doi: 10.1128/mBio.00429-12. mBio. 2012. PMID: 23170000 Free PMC article.
-
Viral Membrane Fusion and the Transmembrane Domain.Viruses. 2020 Jun 27;12(7):693. doi: 10.3390/v12070693. Viruses. 2020. PMID: 32604992 Free PMC article. Review.
-
Residues within the C-terminal arm of the herpes simplex virus 1 glycoprotein B ectodomain contribute to its refolding during the fusion step of virus entry.J Virol. 2012 Jun;86(12):6386-93. doi: 10.1128/JVI.00104-12. Epub 2012 Apr 4. J Virol. 2012. PMID: 22491468 Free PMC article.
-
Silver nanoparticles as potential antiviral agents.Molecules. 2011 Oct 24;16(10):8894-918. doi: 10.3390/molecules16108894. Molecules. 2011. PMID: 22024958 Free PMC article. Review.
-
Membrane fusion, potential threats, and natural antiviral drugs of pseudorabies virus.Vet Res. 2023 May 2;54(1):39. doi: 10.1186/s13567-023-01171-z. Vet Res. 2023. PMID: 37131259 Free PMC article. Review.
References
-
- Adams, P. D., R. W. Grosse-Kunstleve, L. W. Hung, T. R. Ioerger, A. J. McCoy, N. W. Moriarty, R. J. Read, J. C. Sacchettini, N. K. Sauter, and T. C. Terwilliger. 2002. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 58:1948-1954. - PubMed
-
- Atanasiu, D., J. C. Whitbeck, M. P. de Leon, H. Lou, B. P. Hannah, G. H. Cohen, and R. J. Eisenberg. 2010. Bimolecular complementation defines functional regions of herpes simplex virus gB that are involved with gH/gL as a necessary step leading to cell fusion. J. Virol. 84:3825-3834. - PMC - PubMed
-
- Bas, D. C., D. M. Rogers, and J. H. Jensen. 2008. Very fast prediction and rationalization of pKa values for protein-ligand complexes. Proteins 73:765-783. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

