System-level analysis of genes and functions affecting survival during nutrient starvation in Saccharomyces cerevisiae
- PMID: 20944018
- PMCID: PMC3018308
- DOI: 10.1534/genetics.110.120766
System-level analysis of genes and functions affecting survival during nutrient starvation in Saccharomyces cerevisiae
Abstract
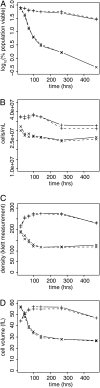
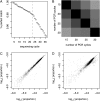
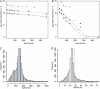
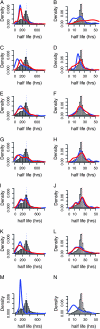
An essential property of all cells is the ability to exit from active cell division and persist in a quiescent state. For single-celled microbes this primarily occurs in response to nutrient deprivation. We studied the genetic requirements for survival of Saccharomyces cerevisiae when starved for either of two nutrients: phosphate or leucine. We measured the survival of nearly all nonessential haploid null yeast mutants in mixed populations using a quantitative sequencing method that estimates the abundance of each mutant on the basis of frequency of unique molecular barcodes. Starvation for phosphate results in a population half-life of 337 hr whereas starvation for leucine results in a half-life of 27.7 hr. To measure survival of individual mutants in each population we developed a statistical framework that accounts for the multiple sources of experimental variation. From the identities of the genes in which mutations strongly affect survival, we identify genetic evidence for several cellular processes affecting survival during nutrient starvation, including autophagy, chromatin remodeling, mRNA processing, and cytoskeleton function. In addition, we found evidence that mitochondrial and peroxisome function is required for survival. Our experimental and analytical methods represent an efficient and quantitative approach to characterizing genetic functions and networks with unprecedented resolution and identified genotype-by-environment interactions that have important implications for interpretation of studies of aging and quiescence in yeast.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

