The structure of SSO2064, the first representative of Pfam family PF01796, reveals a novel two-domain zinc-ribbon OB-fold architecture with a potential acyl-CoA-binding role
- PMID: 20944206
- PMCID: PMC2954200
- DOI: 10.1107/S1744309110002514
The structure of SSO2064, the first representative of Pfam family PF01796, reveals a novel two-domain zinc-ribbon OB-fold architecture with a potential acyl-CoA-binding role
Abstract
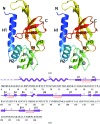
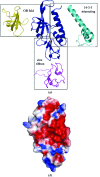
SSO2064 is the first structural representative of PF01796 (DUF35), a large prokaryotic family with a wide phylogenetic distribution. The structure reveals a novel two-domain architecture comprising an N-terminal, rubredoxin-like, zinc ribbon and a C-terminal, oligonucleotide/oligosaccharide-binding (OB) fold domain. Additional N-terminal helical segments may be involved in protein-protein interactions. Domain architectures, genomic context analysis and functional evidence from certain bacterial representatives of this family suggest that these proteins form a novel fatty-acid-binding component that is involved in the biosynthesis of lipids and polyketide antibiotics and that they possibly function as acyl-CoA-binding proteins. This structure has led to a re-evaluation of the DUF35 family, which has now been split into two entries in the latest Pfam release (v.24.0).
Figures

References
Publication types
MeSH terms
Substances
Associated data
- Actions

