Structural classification of proteins and structural genomics: new insights into protein folding and evolution
- PMID: 20944210
- PMCID: PMC2954204
- DOI: 10.1107/S1744309110007177
Structural classification of proteins and structural genomics: new insights into protein folding and evolution
Abstract
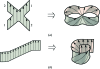
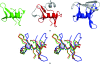
During the past decade, the Protein Structure Initiative (PSI) centres have become major contributors of new families, superfamilies and folds to the Structural Classification of Proteins (SCOP) database. The PSI results have increased the diversity of protein structural space and accelerated our understanding of it. This review article surveys a selection of protein structures determined by the Joint Center for Structural Genomics (JCSG). It presents previously undescribed β-sheet architectures such as the double barrel and spiral β-roll and discusses new examples of unusual topologies and peculiar structural features observed in proteins characterized by the JCSG and other Structural Genomics centres.
Figures

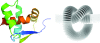
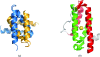


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

