Structure of an essential bacterial protein YeaZ (TM0874) from Thermotoga maritima at 2.5 Å resolution
- PMID: 20944216
- PMCID: PMC2954210
- DOI: 10.1107/S1744309109022192
Structure of an essential bacterial protein YeaZ (TM0874) from Thermotoga maritima at 2.5 Å resolution
Abstract
YeaZ is involved in a protein network that is essential for bacteria. The crystal structure of YeaZ from Thermotoga maritima was determined to 2.5 Å resolution. Although this protein belongs to a family of ancient actin-like ATPases, it appears that it has lost the ability to bind ATP since it lacks some key structural features that are important for interaction with ATP. A conserved surface was identified, supporting its role in the formation of protein complexes.
Figures

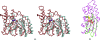
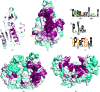
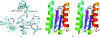
References
-
- Bricogne, G., Vonrhein, C., Flensburg, C., Schiltz, M. & Paciorek, W. (2003). Acta Cryst. D59, 2023–2030. - PubMed
-
- Butland, G., Peregrin-Alvarez, J. M., Li, J., Yang, W., Yang, X., Canadien, V., Starostine, A., Richards, D., Beattie, B., Krogan, N., Davey, M., Parkinson, J., Greenblatt, J. & Emili, A. (2005). Nature (London), 433, 531–537. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

