Structure of a tryptophanyl-tRNA synthetase containing an iron-sulfur cluster
- PMID: 20944229
- PMCID: PMC2954223
- DOI: 10.1107/S1744309110037619
Structure of a tryptophanyl-tRNA synthetase containing an iron-sulfur cluster
Abstract
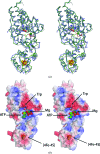
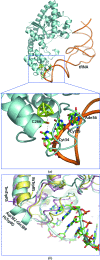
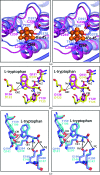
A novel aminoacyl-tRNA synthetase that contains an iron-sulfur cluster in the tRNA anticodon-binding region and efficiently charges tRNA with tryptophan has been found in Thermotoga maritima. The crystal structure of TmTrpRS (tryptophanyl-tRNA synthetase; TrpRS; EC 6.1.1.2) reveals an iron-sulfur [4Fe-4S] cluster bound to the tRNA anticodon-binding (TAB) domain and an L-tryptophan ligand in the active site. None of the other T. maritima aminoacyl-tRNA synthetases (AARSs) contain this [4Fe-4S] cluster-binding motif (C-x₂₂-C-x₆-C-x₂-C). It is speculated that the iron-sulfur cluster contributes to the stability of TmTrpRS and could play a role in the recognition of the anticodon.
Figures



References
Publication types
MeSH terms
Substances
Associated data
- Actions

