The Ndc80 kinetochore complex forms oligomeric arrays along microtubules
- PMID: 20944740
- PMCID: PMC2957311
- DOI: 10.1038/nature09423
The Ndc80 kinetochore complex forms oligomeric arrays along microtubules
Abstract
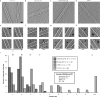
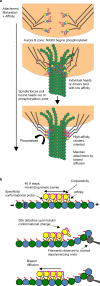
The Ndc80 complex is a key site of regulated kinetochore-microtubule attachment (a process required for cell division), but the molecular mechanism underlying its function remains unknown. Here we present a subnanometre-resolution cryo-electron microscopy reconstruction of the human Ndc80 complex bound to microtubules, sufficient for precise docking of crystal structures of the component proteins. We find that the Ndc80 complex binds the microtubule with a tubulin monomer repeat, recognizing α- and β-tubulin at both intra- and inter-tubulin dimer interfaces in a manner that is sensitive to tubulin conformation. Furthermore, Ndc80 complexes self-associate along protofilaments through interactions mediated by the amino-terminal tail of the NDC80 protein, which is the site of phospho-regulation by Aurora B kinase. The complex's mode of interaction with the microtubule and its oligomerization suggest a mechanism by which Aurora B could regulate the stability of load-bearing kinetochore-microtubule attachments.
Figures


References
-
- Cheeseman IM, Chappie JS, Wilson-Kubalek EM, Desai A. The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell. 2006;127:983–997. - PubMed
-
- Wei RR, Al-Bassam J, Harrison SC. The Ndc80/HEC1 complex is a contact point for kinetochore-microtubule attachment. Nat Struct Mol Biol. 2007;14:54–59. - PubMed
-
- DeLuca JG, et al. Kinetochore microtubule dynamics and attachment stability are regulated by Hec1. Cell. 2006;127:969–982. - PubMed
-
- Martin-Lluesma S, Stucke VM, Nigg EA. Role of Hec1 in spindle checkpoint signaling and kinetochore recruitment of Mad1/Mad2. Science. 2002;297:2267–2270. - PubMed
-
- DeLuca JG, et al. Nuf2 and Hec1 are required for retention of the checkpoint proteins Mad1 and Mad2 to kinetochores. Curr Biol. 2003;13:2103–2109. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

