Structural and thermodynamic analysis of the GFP:GFP-nanobody complex
- PMID: 20945358
- PMCID: PMC3009406
- DOI: 10.1002/pro.519
Structural and thermodynamic analysis of the GFP:GFP-nanobody complex
Abstract
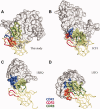
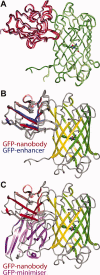
The green fluorescent protein (GFP)-nanobody is a single-chain VHH antibody domain developed with specific binding activity against GFP and is emerging as a powerful tool for isolation and cellular engineering of fluorescent protein fusions in many different fields of biological research. Using X-ray crystallography and isothermal titration calorimetry, we determine the molecular details of GFP:GFP-nanobody complex formation and explain the basis of high affinity and at the same time high specificity of protein binding. Although the GFP-nanobody can also bind YFP, it cannot bind the closely related CFP or other fluorescent proteins from the mFruit series. CFP differs from GFP only within the central chromophore and at one surface amino acid position, which lies in the binding interface. Using this information, we have engineered a CFP variant (I146N) that is also able to bind the GFP-nanobody with high affinity, thus extending the toolbox of genetically encoded fluorescent probes that can be isolated using the GFP-nanobody.
Copyright © 2010 The Protein Society.
Figures

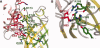


References
-
- Tsien RY. Constructing and exploiting the fluorescent protein paintbox (Nobel Lecture) Angew Chem Int Ed Engl. 2009;48:5612–5626. - PubMed
-
- Skerra A. Alternative non-antibody scaffolds for molecular recognition. Curr Opin Biotechnol. 2007;18:295–304. - PubMed
-
- Muyldermans S, Baral TN, Retamozzo VC, De Baetselier P, De Genst E, Kinne J, Leonhardt H, Magez S, Nguyen VK, Revets H, Rothbauer U, Stijlemans B, Tillib S, Wernery U, Wyns L, Hassanzadeh-Ghassabeh G, Saerens D. Camelid immunoglobulins and nanobody technology. Vet Immunol Immunopathol. 2009;128:178–183. - PubMed
-
- Kirchhofer A, Helma J, Schmidthals K, Frauer C, Cui S, Karcher A, Pellis M, Muyldermans S, Casas-Delucchi CS, Cardoso MC, Leonhardt H, Hopfner KP, Rothbauer U. Modulation of protein properties in living cells using nanobodies. Nat Struct Mol Biol. 2010;17:133–138. - PubMed
-
- Rothbauer U, Zolghadr K, Muyldermans S, Schepers A, Cardoso MC, Leonhardt H. A versatile nanotrap for biochemical and functional studies with fluorescent fusion proteins. Mol Cell Proteomics. 2008;7:282–289. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

