Embryonal carcinoma cell induction of miRNA and mRNA changes in co-cultured prostate stromal fibromuscular cells
- PMID: 20945389
- PMCID: PMC3968429
- DOI: 10.1002/jcp.22464
Embryonal carcinoma cell induction of miRNA and mRNA changes in co-cultured prostate stromal fibromuscular cells
Abstract
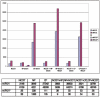
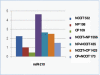
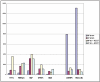
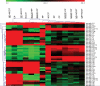
The prostate stromal mesenchyme controls organ-specific development. In cancer, the stromal compartment shows altered gene expression compared to non-cancer. The lineage relationship between cancer-associated stromal cells and normal tissue stromal cells is not known. Nor is the cause underlying the expression difference. Previously, the embryonal carcinoma (EC) cell line, NCCIT, was used by us to study the stromal induction property. In the current study, stromal cells from non-cancer (NP) and cancer (CP) were isolated from tissue specimens and co-cultured with NCCIT cells in a trans-well format to preclude heterotypic cell contact. After 3 days, the stromal cells were analyzed by gene arrays for microRNA (miRNA) and mRNA expression. In co-culture, NCCIT cells were found to alter the miRNA and mRNA expression of NP stromal cells to one like that of CP stromal cells. In contrast, NCCIT had no significant effect on the gene expression of CP stromal cells. We conclude that the gene expression changes in stromal cells can be induced by diffusible factors synthesized by EC cells, and suggest that cancer-associated stromal cells represent a more primitive or less differentiated stromal cell type.
Copyright © 2010 Wiley-Liss, Inc.
Figures





References
-
- Aboseif S, El-Sakka A, Young P, Cunha GR. Mesenchymal reprogramming of adult human epithelial differentiation. Differentiation. 1999;65:113–118. - PubMed
-
- Barroso-del Jesus A, Lucena-Aguilar G, Menendez P. The miR-302-367 cluster as a potential stemness regulator in ESCs. Cell Cycle. 2009;8:394–398. - PubMed
-
- Bonkhoff H, Remberger K. Differentiation pathways and histogenetic aspects of normal and abnormal prostatic growth: a stem cell model. Prostate. 1996;28:98–106. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

