ppGpp is the major source of growth rate control in E. coli
- PMID: 20946586
- PMCID: PMC4556285
- DOI: 10.1111/j.1462-2920.2010.02357.x
ppGpp is the major source of growth rate control in E. coli
Abstract
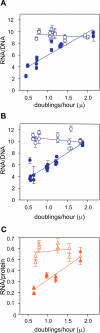
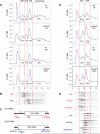
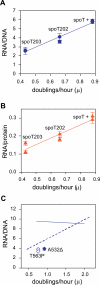
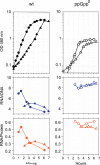
It is widely accepted that the DNA, RNA and protein content of Enterobacteriaceae is regulated as a function of exponential growth rates; macromolecular content increases with faster growth regardless of specific composition of the growth medium. This phenomenon, called growth rate control, primarily involves regulation of ribosomal RNA and ribosomal protein synthesis. However, it was uncertain whether the global regulator ppGpp is the major determinant for growth rate control. Therefore, here we re-evaluate the effect of ppGpp on macromolecular content for different balanced growth rates in defined media. We find that when ppGpp is absent, RNA/protein and RNA/DNA ratios are equivalent in fast and slow growing cells. Moreover, slow growing ppGpp-deficient cells with increased RNA content, display a normal ribosomal subunit composition although polysome content is reduced when compared with fast growing wild-type cells. From this we conclude that growth rate control does not occur in the absence of ppGpp. Also, artificial elevation of ppGpp or introduction of stringent RNA polymerase mutants in ppGpp-deficient cells restores this control. We believe these findings strongly argue in favour of ppGpp and against redundant regulation of growth rate control by other factors in Escherichia coli and other enteric bacteria.
Published 2010. This article is a US Government work and is in the public domain in the USA.
Figures
References
-
- Aberg A, Shingler V, Balsalobre C. Regulation of the fimB promoter: a case of differential regulation by ppGpp and DksA in vivo. Mol Microbiol. 2008;67:1223–1241. - PubMed
-
- Baracchini E, Bremer H. Control of rRNA synthesis in Escherichia coli at increased rrn gene dosage. Role of guanosine tetraphosphate and ribosome feedback. J Biol Chem. 1991;266:11753–11760. - PubMed
-
- Bugl H, Fauman EB, Staker BL, Zheng F, Kushner SR, Saper MA, Bardwell JCA, Jakob U. RNA methylation under heat shock control. Mol Cell. 2000;6:349–360. - PubMed
-
- Bremer H, Dennis PP. Modulation of chemical composition and other parameters of the cell by growth rate. In: Neidhardt FC, editor. Escherichia coli and Salmonella: cellular and molecular biology. ASM Press; Washington DC: 1996. pp. 1553–1569.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

