GOModeler--a tool for hypothesis-testing of functional genomics datasets
- PMID: 20946613
- PMCID: PMC3026376
- DOI: 10.1186/1471-2105-11-S6-S29
GOModeler--a tool for hypothesis-testing of functional genomics datasets
Abstract
Background: Functional genomics technologies that measure genome expression at a global scale are accelerating biological knowledge discovery. Generating these high throughput datasets is relatively easy compared to the downstream functional modelling necessary for elucidating the molecular mechanisms that govern the biology under investigation. A number of publicly available 'discovery-based' computational tools use the computationally amenable Gene Ontology (GO) for hypothesis generation. However, there are few tools that support hypothesis-based testing using the GO and none that support testing with user defined hypothesis terms.Here, we present GOModeler, a tool that enables researchers to conduct hypothesis-based testing of high throughput datasets using the GO. GOModeler summarizes the overall effect of a user defined gene/protein differential expression dataset on specific GO hypothesis terms selected by the user to describe a biological experiment. The design of the tool allows the user to complement the functional information in the GO with his/her domain specific expertise for comprehensive hypothesis testing.
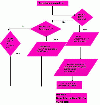
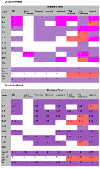
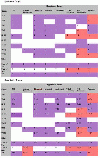
Results: GOModeler tests the relevance of the hypothesis terms chosen by the user for the input gene dataset by providing the individual effects of the genes on the hypothesis terms and the overall effect of the entire dataset on each of the hypothesis terms. It matches the GO identifiers (ids) of the genes with the GO ids of the hypothesis terms and parses the names of those ids that match to assign effects. We demonstrate the capabilities of GOModeler with a dataset of nine differentially expressed cytokine genes and compare the results to those obtained through manual analysis of the dataset by an immunologist. The direction of overall effects on all hypothesis terms except one was consistent with the results obtained by manual analysis. The tool's editing capability enables the user to augment the information extracted. GOModeler is available as a part of the AgBase tool suite (http://www.agbase.msstate.edu).
Conclusions: GOModeler allows hypothesis driven analysis of high throughput datasets using the GO. Using this tool, researchers can quickly evaluate the overall effect of quantitative expression changes of gene set on specific biological processes of interest. The results are provided in both tabular and graphical formats.
Figures






References
MeSH terms
LinkOut - more resources
Full Text Sources

