Assessing constancy of substitution rates in viruses over evolutionary time
- PMID: 20946614
- PMCID: PMC3026377
- DOI: 10.1186/1471-2105-11-S6-S3
Assessing constancy of substitution rates in viruses over evolutionary time
Abstract
Background: Phylogenetic analyses reveal probable patterns of divergence of present day organisms from common ancestors. The points of divergence of lineages can be dated if a corresponding historical or fossil record exists. For many species, in particular viruses, such records are rare. Recently, Bayesian phylogenetic analysis using sequences from closely related organisms isolated at different times have been used to calibrate divergences. Phylogenetic analyses depend on the assumption that the average substitution rates that can be calculated from the data apply throughout the course of evolution.
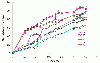
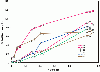
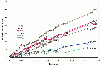
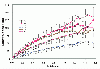
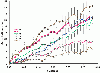
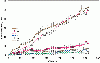
Results: The present study tests this crucial assumption by charting the kinds of substitutions observed between pairs of sequences with different levels of total substitutions. Datasets of aligned sequences, both viral and non-viral, were assembled. For each pair of sequences in an aligned set, the distribution of nucleotide interchanges and the total number of changes were calculated. Data were binned according to total numbers of changes and plotted. The accumulation of the six possible interchange types in retroelements as a function of distance followed closely the expected hyperbolic relationship. For other datasets, however, significant deviations from this relationship were noted. A rapid initial accumulation of transition interchanges was frequent among the datasets and anomalous changes occurred at specific divergence levels.
Conclusions: The accumulation profiles suggested that substantial changes in frequencies of types of substitutions occur over the course of evolution and that such changes should be considered in evaluating and dating viral phylogenies.
Figures
Similar articles
-
Declining transition/transversion ratios through time reveal limitations to the accuracy of nucleotide substitution models.BMC Evol Biol. 2015 Mar 11;15:36. doi: 10.1186/s12862-015-0312-6. BMC Evol Biol. 2015. PMID: 25886870 Free PMC article.
-
Bayesian Molecular Clock Dating Using Genome-Scale Datasets.Methods Mol Biol. 2019;1910:309-330. doi: 10.1007/978-1-4939-9074-0_10. Methods Mol Biol. 2019. PMID: 31278669
-
Angiosperm divergence times: the effect of genes, codon positions, and time constraints.Evolution. 2005 Aug;59(8):1653-70. doi: 10.1554/04-565.1. Evolution. 2005. PMID: 16329238
-
Inference of viral evolutionary rates from molecular sequences.Adv Parasitol. 2003;54:331-58. doi: 10.1016/s0065-308x(03)54008-8. Adv Parasitol. 2003. PMID: 14711090 Review.
-
Ribosomal DNA: molecular evolution and phylogenetic inference.Q Rev Biol. 1991 Dec;66(4):411-53. doi: 10.1086/417338. Q Rev Biol. 1991. PMID: 1784710 Review.
Cited by
-
Proceedings of the 2010 MidSouth Computational Biology and Bioinformatics Society (MCBIOS) conference.BMC Bioinformatics. 2010 Oct 7;11 Suppl 6(Suppl 6):S1. doi: 10.1186/1471-2105-11-S6-S1. BMC Bioinformatics. 2010. PMID: 20946592 Free PMC article. No abstract available.
-
Proceedings of the 2011 MidSouth Computational Biology and Bioinformatics Society (MCBIOS) conference. Introduction.BMC Bioinformatics. 2011 Oct 18;12 Suppl 10(Suppl 10):S1. doi: 10.1186/1471-2105-12-S10-S1. BMC Bioinformatics. 2011. PMID: 22165918 Free PMC article. No abstract available.
References
-
- Lartey RT, Voss TC, Melcher U. Tobamovirus evolution: gene overlaps, recombination, and taxonomic implications. Mol Biol Evol. 1996;13:1327–1338. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

