Systematic identification of conserved motif modules in the human genome
- PMID: 20946653
- PMCID: PMC3091716
- DOI: 10.1186/1471-2164-11-567
Systematic identification of conserved motif modules in the human genome
Abstract
Background: The identification of motif modules, groups of multiple motifs frequently occurring in DNA sequences, is one of the most important tasks necessary for annotating the human genome. Current approaches to identifying motif modules are often restricted to searches within promoter regions or rely on multiple genome alignments. However, the promoter regions only account for a limited number of locations where transcription factor binding sites can occur, and multiple genome alignments often cannot align binding sites with their true counterparts because of the short and degenerative nature of these transcription factor binding sites.
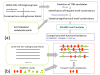
Results: To identify motif modules systematically, we developed a computational method for the entire non-coding regions around human genes that does not rely upon the use of multiple genome alignments. First, we selected orthologous DNA blocks approximately 1-kilobase in length based on discontiguous sequence similarity. Next, we scanned the conserved segments in these blocks using known motifs in the TRANSFAC database. Finally, a frequent pattern mining technique was applied to identify motif modules within these blocks. In total, with a false discovery rate cutoff of 0.05, we predicted 3,161,839 motif modules, 90.8% of which are supported by various forms of functional evidence. Compared with experimental data from 14 ChIP-seq experiments, on average, our methods predicted 69.6% of the ChIP-seq peaks with TFBSs of multiple TFs. Our findings also show that many motif modules have distance preference and order preference among the motifs, which further supports the functionality of these predictions.
Conclusions: Our work provides a large-scale prediction of motif modules in mammals, which will facilitate the understanding of gene regulation in a systematic way.
Figures