The Structure of the talin head reveals a novel extended conformation of the FERM domain
- PMID: 20947018
- PMCID: PMC2977851
- DOI: 10.1016/j.str.2010.07.011
The Structure of the talin head reveals a novel extended conformation of the FERM domain
Abstract
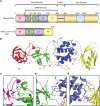
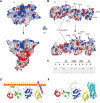
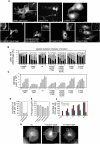
FERM domains are found in a diverse superfamily of signaling and adaptor proteins at membrane interfaces. They typically consist of three separately folded domains (F1, F2, F3) in a compact cloverleaf structure. The crystal structure of the N-terminal head of the integrin-associated cytoskeletal protein talin reported here reveals a novel FERM domain with a linear domain arrangement, plus an additional domain F0 packed against F1. While F3 binds β-integrin tails, basic residues in F1 and F2 are required for membrane association and for integrin activation. We show that these same residues are also required for cell spreading and focal adhesion assembly in cells. We suggest that the extended conformation of the talin head allows simultaneous binding to integrins via F3 and to PtdIns(4,5)P2-enriched microdomains via basic residues distributed along one surface of the talin head, and that these multiple interactions are required to stabilize integrins in the activated state.
Copyright © 2010 Elsevier Ltd. All rights reserved.
Figures



Comment in
-
The talin FERM domain is not so FERM.Structure. 2010 Oct 13;18(10):1222-3. doi: 10.1016/j.str.2010.09.002. Structure. 2010. PMID: 20947007
References
-
- Barsukov I.L., Prescot A., Bate N., Patel B., Floyd D.N., Bhanji N., Bagshaw C.R., Lethinic K., Di Paolo G., De Camilli P. Phosphatidylinositol phosphate kinase type 1gamma and beta1-integrin cytoplasmic domain bind to the same region in the talin FERM domain. J. Biol. Chem. 2003;278:31202–31209. - PubMed
-
- Bernado P., Mylonas E., Petoukhov M.V., Blackledge M., Svergun D.I. Structural characterization of flexible proteins using small-angle X-ray scattering. J. Am. Chem. Soc. 2007;129:5656–5664. - PubMed
-
- Bouaouina M., Lad Y., Calderwood D.A. The N-terminal domains of talin cooperate with the phosphotyrosine binding-like domain to activate β1 and β3 integrins. J. Biol. Chem. 2008;283:6118–6125. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

