Hydrogen/deuterium exchange reveals distinct agonist/partial agonist receptor dynamics within vitamin D receptor/retinoid X receptor heterodimer
- PMID: 20947021
- PMCID: PMC2956612
- DOI: 10.1016/j.str.2010.07.007
Hydrogen/deuterium exchange reveals distinct agonist/partial agonist receptor dynamics within vitamin D receptor/retinoid X receptor heterodimer
Abstract
Regulation of nuclear receptor (NR) activity is driven by alterations in the conformational dynamics of the receptor upon ligand binding. Previously, we demonstrated that hydrogen/deuterium exchange (HDX) can be applied to determine novel mechanism of action of PPARγ ligands and in predicting tissue specificity of selective estrogen receptor modulators. Here, we applied HDX to probe the conformational dynamics of the ligand binding domain (LBD) of the vitamin D receptor (VDR) upon binding its natural ligand 1α,25-dihydroxyvitamin D3 (1,25D3), and two analogs, alfacalcidol and ED-71. Comparison of HDX profiles from ligands in complex with the LBD with full-length receptor bound to its cognate receptor retinoid X receptor (RXR) revealed unique receptor dynamics that could not be inferred from static crystal structures. These results demonstrate that ligands modulate the dynamics of the heterodimer interface as well as provide insight into the role of AF-2 dynamics in the action of VDR partial agonists.
Copyright © 2010 Elsevier Ltd. All rights reserved.
Figures
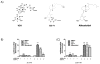


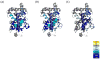
Comment in
-
Dynamics of nuclear receptors.Structure. 2010 Oct 13;18(10):1225-7. doi: 10.1016/j.str.2010.09.003. Structure. 2010. PMID: 20947009
References
-
- Bruning JB, Chalmers MJ, Prasad S, Busby SA, Kamenecka TM, He Y, Nettles KW, Griffin PR. Partial agonists activate PPARgamma using a helix 12 independent mechanism. Structure. 2007;15:1258–1271. - PubMed
-
- Chalmers MJ, Busby SA, Pascal BD, He Y, Hendrickson CL, Marshall AG, Griffin PR. Probing protein ligand interactions by automated hydrogen/deuterium exchange mass spectrometry. Anal Chem. 2006;78:1005–1014. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

