Fluorescent xDNA nucleotides as efficient substrates for a template-independent polymerase
- PMID: 20947563
- PMCID: PMC3045586
- DOI: 10.1093/nar/gkq853
Fluorescent xDNA nucleotides as efficient substrates for a template-independent polymerase
Abstract
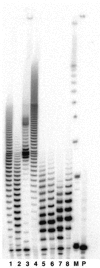
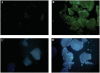
Template independent polymerases, and terminal deoxynucleotidyl transferase (TdT) in particular, have been widely used in enzymatic labeling of DNA 3'-ends, yielding fluorescently-labeled polymers. The majority of fluorescent nucleotides used as TdT substrates contain tethered fluorophores attached to a natural nucleotide, and can be hindered by undesired fluorescence characteristics such as self-quenching. We previously documented the inherent fluorescence of a set of four benzo-expanded deoxynucleoside analogs (xDNA) that maintain Watson-Crick base pairing and base stacking ability; however, their substrate abilities for standard template-dependent polymerases were hampered by their large size. However, it seemed possible that a template-independent enzyme, due to lowered geometric constraints, might be less restrictive of nucleobase size. Here, we report the synthesis and study of xDNA nucleoside triphosphates, and studies of their substrate abilities with TdT. We find that this polymerase can incorporate each of the four xDNA monomers with kinetic efficiencies that are nearly the same as those of natural nucleotides, as measured by steady-state methods. As many as 30 consecutive monomers could be incorporated. Fluorescence changes over time could be observed in solution during the enzymatic incorporation of expanded adenine (dxATP) and cytosine (dxCTP) analogs, and after incorporation, when attached to a glass solid support. For (dxA)(n) polymers, monomer emission quenching and long-wavelength excimer emission was observed. For (dxC)(n), fluorescence enhancement was observed in the polymer. TdT-mediated synthesis may be a useful approach for creating xDNA labels or tags on DNA, making use of the fluorescence and strong hybridization properties of the xDNA.
Figures
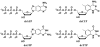
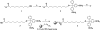

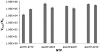

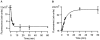
Similar articles
-
Stereoisomers of deoxynucleoside 5'-triphosphates as substrates for template-dependent and -independent DNA polymerases.J Biol Chem. 1997 Apr 4;272(14):9556-60. doi: 10.1074/jbc.272.14.9556. J Biol Chem. 1997. PMID: 9083099
-
Computational Modeling Study of the Molecular Basis of dNTP Selectivity in Human Terminal Deoxynucleotidyltransferase.Biomolecules. 2024 Aug 7;14(8):961. doi: 10.3390/biom14080961. Biomolecules. 2024. PMID: 39199349 Free PMC article.
-
Pre-steady-state kinetic studies of the fidelity of human DNA polymerase mu.Biochemistry. 2004 Nov 2;43(43):13827-38. doi: 10.1021/bi048782m. Biochemistry. 2004. PMID: 15504045
-
Terminal Deoxynucleotidyl Transferase in the Synthesis and Modification of Nucleic Acids.Chembiochem. 2019 Apr 1;20(7):860-871. doi: 10.1002/cbic.201800658. Epub 2019 Jan 25. Chembiochem. 2019. PMID: 30451377 Review.
-
Mutagenesis by incorporation of alkylated nucleotides.Basic Life Sci. 1985;31:339-51. doi: 10.1007/978-1-4613-2449-2_21. Basic Life Sci. 1985. PMID: 3888179 Review.
Cited by
-
DNA polymerase θ specializes in incorporating synthetic expanded-size (xDNA) nucleotides.Nucleic Acids Res. 2016 Nov 2;44(19):9381-9392. doi: 10.1093/nar/gkw721. Epub 2016 Sep 2. Nucleic Acids Res. 2016. PMID: 27591252 Free PMC article.
-
Template-independent synthesis and 3'-end labelling of 2'-modified oligonucleotides with terminal deoxynucleotidyl transferases.Nucleic Acids Res. 2024 Sep 23;52(17):10085-10101. doi: 10.1093/nar/gkae691. Nucleic Acids Res. 2024. PMID: 39149896 Free PMC article.
-
Modified Nucleotides as Substrates of Terminal Deoxynucleotidyl Transferase.Molecules. 2017 Apr 22;22(4):672. doi: 10.3390/molecules22040672. Molecules. 2017. PMID: 28441732 Free PMC article.
-
Fluorescent DNA-based enzyme sensors.Chem Soc Rev. 2011 Dec;40(12):5756-70. doi: 10.1039/c0cs00162g. Epub 2011 Feb 2. Chem Soc Rev. 2011. PMID: 21290032 Free PMC article. Review.
-
A versatile method for the UVA-induced cross-linking of acetophenone- or benzophenone-functionalized DNA.Sci Rep. 2018 Nov 7;8(1):16484. doi: 10.1038/s41598-018-34892-9. Sci Rep. 2018. PMID: 30405165 Free PMC article.
References
-
- Vineyard D, Zhang X, Donnelly A, Lee I, Berdis AJ. Optimization of non-natural nucleotides for selective incorporation opposite damaged DNA. Org. Biomol. Chem. 2007;5:3623–3630. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

