Rapid construction of empirical RNA fitness landscapes
- PMID: 20947767
- PMCID: PMC3392653
- DOI: 10.1126/science.1192001
Rapid construction of empirical RNA fitness landscapes
Abstract
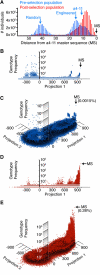
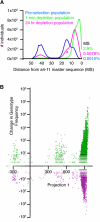
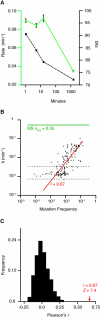
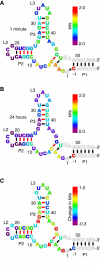
Evolution is an adaptive walk through a hypothetical fitness landscape, which depicts the relationship between genotypes and the fitness of each corresponding phenotype. We constructed an empirical fitness landscape for a catalytic RNA by combining next-generation sequencing, computational analysis, and "serial depletion," an in vitro selection protocol. By determining the reaction rate constant for every point mutant of a catalytic RNA, we demonstrated that abundance in serially depleted pools correlates with biochemical activity (correlation coefficient r = 0.67, standard score Z = 7.4). Therefore, enumeration of each genotype by deep sequencing yielded a fitness landscape containing ~10(7) unique sequences, without requiring measurement of the phenotypic fitness for each sequence. High-throughput mapping between genotype and phenotype may apply to artificial selections, host-pathogen interactions, and other biomedically relevant evolutionary phenomena.
Figures
Comment in
-
Evolution. RNA GPS.Science. 2010 Oct 15;330(6002):330-1. doi: 10.1126/science.1197667. Science. 2010. PMID: 20947753 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

