RB-pathway disruption in breast cancer: differential association with disease subtypes, disease-specific prognosis and therapeutic response
- PMID: 20948315
- PMCID: PMC3055199
- DOI: 10.4161/cc.9.20.13454
RB-pathway disruption in breast cancer: differential association with disease subtypes, disease-specific prognosis and therapeutic response
Abstract
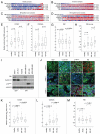
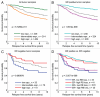
In breast cancer, inactivation of the RB tumor suppressor gene is believed to occur via multiple mechanisms to facilitate tumorigenesis. However, the prognostic and predictive value of RB status in disease-specific clinical outcomes has remained uncertain. We investigated RB pathway deregulation in the context of both ER-positive and ER-negative disease using combined microarray datasets encompassing over 900 breast cancer patient samples. Disease-specific characteristics of RB pathway deregulation were investigated in this dataset by evaluating correlation among pathway genes as well as differential expression across patient tumor populations defined by ER status. Survival analysis among these breast cancer samples demonstrates that the RB-loss signature is associated with poor disease outcome within several independent cohorts. Within the ER-negative subpopulation, the RB-loss signature is associated with improved response to chemotherapy and longer relapse-free survival. Additionally, while individual genes in the RB target signature closely reproduce its prognostic value, they also serve to predict and monitor response to therapeutic compounds, such as the cytostatic agent PD-0332991. These results indicate that the RB-loss signature expression is associated with poor outcome in breast cancer, but predicts improved response to chemotherapy based on data in ER-negative populations. While the RB-loss signature, as a whole, demonstrates prognostic and predictive utility, a small subset of markers could be sufficient to stratify patients based on RB function and inform the selection of appropriate therapeutic regimens.
Figures




Comment in
-
RB in breast cancer: differential effects in estrogen receptor-positive and estrogen receptor-negative disease.Cell Cycle. 2010 Dec 1;9(23):4607. doi: 10.4161/cc.9.23.13889. Cell Cycle. 2010. PMID: 21260944 Free PMC article. No abstract available.
Similar articles
-
RB-pathway disruption is associated with improved response to neoadjuvant chemotherapy in breast cancer.Clin Cancer Res. 2012 Sep 15;18(18):5110-22. doi: 10.1158/1078-0432.CCR-12-0903. Epub 2012 Jul 18. Clin Cancer Res. 2012. PMID: 22811582 Free PMC article.
-
A gene expression signature of retinoblastoma loss-of-function is a predictive biomarker of resistance to palbociclib in breast cancer cell lines and is prognostic in patients with ER positive early breast cancer.Oncotarget. 2016 Sep 13;7(42):68012-68022. doi: 10.18632/oncotarget.12010. Oncotarget. 2016. PMID: 27634906 Free PMC article.
-
Prognostic breast cancer signature identified from 3D culture model accurately predicts clinical outcome across independent datasets.PLoS One. 2008 Aug 20;3(8):e2994. doi: 10.1371/journal.pone.0002994. PLoS One. 2008. PMID: 18714348 Free PMC article.
-
Retinoblastoma tumor suppressor pathway in breast cancer: prognosis, precision medicine, and therapeutic interventions.Breast Cancer Res. 2014 May 7;16(3):207. doi: 10.1186/bcr3652. Breast Cancer Res. 2014. PMID: 25223380 Free PMC article. Review.
-
RB in breast cancer: at the crossroads of tumorigenesis and treatment.Cell Cycle. 2007 Mar 15;6(6):667-71. doi: 10.4161/cc.6.6.3988. Epub 2007 Mar 7. Cell Cycle. 2007. PMID: 17361100 Review.
Cited by
-
Molecular damage in cancer: an argument for mTOR-driven aging.Aging (Albany NY). 2011 Dec;3(12):1130-41. doi: 10.18632/aging.100422. Aging (Albany NY). 2011. PMID: 22246147 Free PMC article.
-
Therapeutic response to CDK4/6 inhibition in breast cancer defined by ex vivo analyses of human tumors.Cell Cycle. 2012 Jul 15;11(14):2756-61. doi: 10.4161/cc.21195. Epub 2012 Jul 15. Cell Cycle. 2012. PMID: 22767154 Free PMC article.
-
RB1: a prototype tumor suppressor and an enigma.Genes Dev. 2016 Jul 1;30(13):1492-502. doi: 10.1101/gad.282145.116. Genes Dev. 2016. PMID: 27401552 Free PMC article. Review.
-
A Human Adult Stem Cell Signature Marks Aggressive Variants across Epithelial Cancers.Cell Rep. 2018 Sep 18;24(12):3353-3366.e5. doi: 10.1016/j.celrep.2018.08.062. Cell Rep. 2018. PMID: 30232014 Free PMC article.
-
Chromatin-bound RB targets promoters, enhancers, and CTCF-bound loci and is redistributed by cell-cycle progression.Mol Cell. 2022 Sep 15;82(18):3333-3349.e9. doi: 10.1016/j.molcel.2022.07.014. Epub 2022 Aug 17. Mol Cell. 2022. PMID: 35981542 Free PMC article.
References
-
- American Cancer Society, author. Cancer facts & figures 2009. Atlanta, GA: The American Cancer Society; 2009.
-
- Musgrove EA, Sutherland RL. Biological determinants of endocrine resistance in breast cancer. Nat Rev Cancer. 2009;9:631–643. - PubMed
-
- Schneider BP, Winer EP, Foulkes WD, Garber J, Perou CM, Richardson A, et al. Triple-negative breast cancer: risk factors to potential targets. Clin Cancer Res. 2008;14:8010–8018. - PubMed
-
- Reis-Filho JS, Tutt AN. Triple negative tumours: a critical review. Histopathology. 2008;52:108–118. - PubMed
-
- Miller TW, Perez-Torres M, Narasanna A, Guix M, Stal O, Perez-Tenorio G, et al. Loss of Phosphatase and Tensin homologue deleted on chromosome 10 engages ErbB3 and insulin-like growth factor-I receptor signaling to promote antiestrogen resistance in breast cancer. Cancer Res. 2009;69:4192–4201. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
