Proteomics: challenges, techniques and possibilities to overcome biological sample complexity
- PMID: 20948568
- PMCID: PMC2950283
- DOI: 10.4061/2009/239204
Proteomics: challenges, techniques and possibilities to overcome biological sample complexity
Abstract
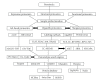
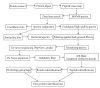
Proteomics is the large-scale study of the structure and function of proteins in complex biological sample. Such an approach has the potential value to understand the complex nature of the organism. Current proteomic tools allow large-scale, high-throughput analyses for the detection, identification, and functional investigation of proteome. Advances in protein fractionation and labeling techniques have improved protein identification to include the least abundant proteins. In addition, proteomics has been complemented by the analysis of posttranslational modifications and techniques for the quantitative comparison of different proteomes. However, the major limitation of proteomic investigations remains the complexity of biological structures and physiological processes, rendering the path of exploration paved with various difficulties and pitfalls. The quantity of data that is acquired with new techniques places new challenges on data processing and analysis. This article provides a brief overview of currently available proteomic techniques and their applications, followed by detailed description of advantages and technical challenges. Some solutions to circumvent technical difficulties are proposed.
Figures




References
-
- Wilkins MR, Sanchez J-C, Gooley AA, et al. Progress with proteome projects: why all proteins expressed by a genome should be identified and how to do it. Biotechnology and Genetic Engineering Reviews. 1995;13:19–50. - PubMed
-
- Baltimore D. Our genome unveiled. Nature. 2001;409(6822):814–816. - PubMed
-
- Wilkins MR, Gasteiger E, Gooley AA, et al. High-throughput mass spectrometric discovery of protein post-translational modifications. Journal of Molecular Biology. 1999;289(3):645–657. - PubMed
-
- Godovac-Zimmermann J, Brown LR. Perspectives for mass spectrumetry and functional proteomics. Mass Spectrometry Reviews. 2001;20(1):1–57. - PubMed
LinkOut - more resources
Full Text Sources
