Staphylococcus epidermidis strategies to avoid killing by human neutrophils
- PMID: 20949069
- PMCID: PMC2951371
- DOI: 10.1371/journal.ppat.1001133
Staphylococcus epidermidis strategies to avoid killing by human neutrophils
Abstract
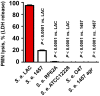
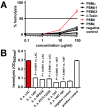
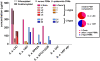
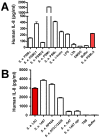
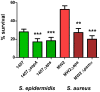
Staphylococcus epidermidis is a leading nosocomial pathogen. In contrast to its more aggressive relative S. aureus, it causes chronic rather than acute infections. In highly virulent S. aureus, phenol-soluble modulins (PSMs) contribute significantly to immune evasion and aggressive virulence by their strong ability to lyse human neutrophils. Members of the PSM family are also produced by S. epidermidis, but their role in immune evasion is not known. Notably, strong cytolytic capacity of S. epidermidis PSMs would be at odds with the notion that S. epidermidis is a less aggressive pathogen than S. aureus, prompting us to examine the biological activities of S. epidermidis PSMs. Surprisingly, we found that S. epidermidis has the capacity to produce PSMδ, a potent leukocyte toxin, representing the first potent cytolysin to be identified in that pathogen. However, production of strongly cytolytic PSMs was low in S. epidermidis, explaining its low cytolytic potency. Interestingly, the different approaches of S. epidermidis and S. aureus to causing human disease are thus reflected by the adaptation of biological activities within one family of virulence determinants, the PSMs. Nevertheless, S. epidermidis has the capacity to evade neutrophil killing, a phenomenon we found is partly mediated by resistance mechanisms to antimicrobial peptides (AMPs), including the protease SepA, which degrades AMPs, and the AMP sensor/resistance regulator, Aps (GraRS). These findings establish a significant function of SepA and Aps in S. epidermidis immune evasion and explain in part why S. epidermidis may evade elimination by innate host defense despite the lack of cytolytic toxin expression. Our study shows that the strategy of S. epidermidis to evade elimination by human neutrophils is characterized by a passive defense approach and provides molecular evidence to support the notion that S. epidermidis is a less aggressive pathogen than S. aureus.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



Similar articles
-
Phenol-Soluble Modulin Toxins of Staphylococcus haemolyticus.Front Cell Infect Microbiol. 2017 May 24;7:206. doi: 10.3389/fcimb.2017.00206. eCollection 2017. Front Cell Infect Microbiol. 2017. PMID: 28596942 Free PMC article.
-
The virulence regulator Agr controls the staphylococcal capacity to activate human neutrophils via the formyl peptide receptor 2.J Innate Immun. 2012;4(2):201-12. doi: 10.1159/000332142. Epub 2011 Nov 8. J Innate Immun. 2012. PMID: 22067547 Free PMC article.
-
Staphylococcus aureus Depends on Eap Proteins for Preventing Degradation of Its Phenol-Soluble Modulin Toxins by Neutrophil Serine Proteases.Front Immunol. 2021 Sep 6;12:701093. doi: 10.3389/fimmu.2021.701093. eCollection 2021. Front Immunol. 2021. PMID: 34552584 Free PMC article.
-
Molecular basis of Staphylococcus epidermidis infections.Semin Immunopathol. 2012 Mar;34(2):201-14. doi: 10.1007/s00281-011-0296-2. Epub 2011 Nov 19. Semin Immunopathol. 2012. PMID: 22095240 Free PMC article. Review.
-
Phenol-soluble modulins: novel virulence-associated peptides of staphylococci.Future Microbiol. 2014;9(2):203-16. doi: 10.2217/fmb.13.153. Epub 2013 Dec 3. Future Microbiol. 2014. PMID: 24295365 Review.
Cited by
-
Clinical MRSA isolates from skin and soft tissue infections show increased in vitro production of phenol soluble modulins.J Infect. 2015 Oct;71(4):447-57. doi: 10.1016/j.jinf.2015.06.005. Epub 2015 Jun 14. J Infect. 2015. PMID: 26079275 Free PMC article.
-
The road to success of coagulase-negative staphylococci: clinical significance of small colony variants and their pathogenic role in persistent infections.Eur J Clin Microbiol Infect Dis. 2021 Nov;40(11):2249-2270. doi: 10.1007/s10096-021-04315-1. Epub 2021 Jul 23. Eur J Clin Microbiol Infect Dis. 2021. PMID: 34296355 Free PMC article. Review.
-
Bacterial Evasion of Host Antimicrobial Peptide Defenses.Microbiol Spectr. 2016 Feb;4(1):10.1128/microbiolspec.VMBF-0006-2015. doi: 10.1128/microbiolspec.VMBF-0006-2015. Microbiol Spectr. 2016. PMID: 26999396 Free PMC article. Review.
-
Phenol-Soluble Modulin Toxins of Staphylococcus haemolyticus.Front Cell Infect Microbiol. 2017 May 24;7:206. doi: 10.3389/fcimb.2017.00206. eCollection 2017. Front Cell Infect Microbiol. 2017. PMID: 28596942 Free PMC article.
-
The Skin Microbiota: Balancing Risk and Reward.Cell Host Microbe. 2020 Aug 12;28(2):190-200. doi: 10.1016/j.chom.2020.06.017. Cell Host Microbe. 2020. PMID: 32791112 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

