Characterization and comparison of two binding sites on obscurin for small ankyrin 1
- PMID: 20949908
- PMCID: PMC3000613
- DOI: 10.1021/bi101165p
Characterization and comparison of two binding sites on obscurin for small ankyrin 1
Abstract
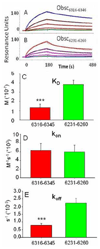
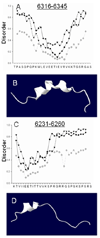
Obscurin A, an ∼720 kDa modular protein of striated muscles, binds to small ankyrin 1 (sAnk1, Ank 1.5), an integral protein of the sarcoplasmic reticulum, through two distinct carboxy-terminal sequences, Obsc(6316-6436) and Obsc(6236-6260). We hypothesized that these sequences differ in affinity but that they compete for the same binding site on sAnk1. We show that the sequence within Obsc(6316-6436) that binds to sAnk1 is limited to residues 6316-6345. Comparison of Obsc(6231-6260) to Obsc(6316-6345) reveals that Obsc(6316-6345) binds sAnk1 with an affinity (133 ± 43 nM) comparable to that of the Obsc(6316-6436) fusion protein, whereas Obsc(6231-6260) binds with lower affinity (384 ± 53 nM). Oligopeptides of each sequence compete for binding with both sites at half-maximal inhibitory concentrations consistent with the affinities measured directly. Five of six site-directed mutants of sAnk1 showed similar reductions in binding to each binding site on obscurin, suggesting that they dock to many of the same residues of sAnk1. Circular dichroism (CD) analysis of the synthetic oligopeptides revealed a 2-fold greater α-helical content in Obsc(6316-6346), ∼35%, than Obsc(6231-6260,) ∼17%. Using these data, structural prediction algorithms, and homology modeling, we predict that Obsc(6316-6345) contains a bent α-helix of 12 amino acids, flanked by short disordered regions, and that Obsc(6231-6260) has a short, N-terminal α-helix of 4-5 residues followed by a long disordered region. Our results are consistent with a model in which both sequences of obscurin differ significantly in structure but bind to the ankyrin-like repeat motifs of sAnk1 in a similar though not identical manner.
Figures




References
-
- Birkenmeier CS, Sharp JJ, Gifford EJ, Deveau SA, Barker JE. An alternative first exon in the distal end of the erythroid ankyrin gene leads to production of a small isoform containing an NH2-terminal membrane anchor. Genomics. 1998;50:79–88. - PubMed
-
- Bennett V, Baines AJ. Spectrin and ankyrin-based pathways: metazoan inventions for integrating cells into tissues. Physiol Rev. 2001;81:1353–1392. - PubMed
-
- Carlsson L, Yu JG, Thornell LE. New aspects of obscurin in human striated muscles. Histochem. Cell Biol. 2008;130:91–103. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

