Planctomycetes dominate biofilms on surfaces of the kelp Laminaria hyperborea
- PMID: 20950420
- PMCID: PMC2964680
- DOI: 10.1186/1471-2180-10-261
Planctomycetes dominate biofilms on surfaces of the kelp Laminaria hyperborea
Abstract
Background: Bacteria belonging to Planctomycetes display several unique morphological and genetic features and are found in a wide variety of habitats on earth. Their ecological roles in these habitats are still poorly understood. Planctomycetes have previously been detected throughout the year on surfaces of the kelp Laminaria hyperborea from southwestern Norway. We aimed to make a detailed investigation of the abundance and phylogenetic diversity of planctomycetes inhabiting these kelp surfaces.
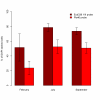
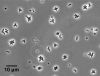
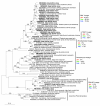
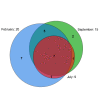
Results: Planctomycetes accounted for 51-53% of the bacterial biofilm cells in July and September and 24% in February according to fluorescence in situ hybridization (FISH) results. Several separate planctomycetes lineages within Pirellulae, Planctomyces and OM190 were represented in 16S rRNA gene clone libraries and the most abundant clones belonged to yet uncultured lineages. In contrast to the abundance, the diversity of the planctomycete populations increased from July to February and was probably influenced by the aging of the kelp tissue. One planctomycete strain that was closely related to Rhodopirellula baltica was isolated using selective cultivation techniques.
Conclusions: Biofilms on surfaces of L. hyperborea display an even higher proportion of planctomycetes compared to other investigated planctomycete-rich habitats such as open water, sandy sediments and peat bogs. The findings agree well with the hypothesis of the role of planctomycetes as degraders of sulfated polymeric carbon in the marine environment as kelps produce such substances. In addition, the abundant planctomycete populations on kelp surfaces and in association with other eukaryotes suggest that coexistence with eukaryotes may be a key feature of many planctomycete lifestyles.
Figures

References
-
- Ward N, Staley JT, Fuerst JA, Giovannoni S, Schlesner H, Stackebrandt E. The Prokaryotes. 3. Vol. 7. New York: Springer; 2006. The Order Planctomycetales, Including the Genera Planctomyces, Pirellula, Gemmata and Isosphaera and the Candidatus Genera Brocadia, Kuenenia and Scalindua.
-
- Delong EF, Franks DG, Alldredge AL. Phylogenetic diversity of aggregate-attached vs. free-living marine bacterial assemblages. Limnol Oceanogr. 1993;38(5):924–934. doi: 10.4319/lo.1993.38.5.0924. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

