Genome-wide characterization of simple sequence repeats in cucumber (Cucumis sativus L.)
- PMID: 20950470
- PMCID: PMC3091718
- DOI: 10.1186/1471-2164-11-569
Genome-wide characterization of simple sequence repeats in cucumber (Cucumis sativus L.)
Abstract
Background: Cucumber, Cucumis sativus L. is an important vegetable crop worldwide. Until very recently, cucumber genetic and genomic resources, especially molecular markers, have been very limited, impeding progress of cucumber breeding efforts. Microsatellites are short tandemly repeated DNA sequences, which are frequently favored as genetic markers due to their high level of polymorphism and codominant inheritance. Data from previously characterized genomes has shown that these repeats vary in frequency, motif sequence, and genomic location across taxa. During the last year, the genomes of two cucumber genotypes were sequenced including the Chinese fresh market type inbred line '9930' and the North American pickling type inbred line 'Gy14'. These sequences provide a powerful tool for developing markers in a large scale. In this study, we surveyed and characterized the distribution and frequency of perfect microsatellites in 203 Mbp assembled Gy14 DNA sequences, representing 55% of its nuclear genome, and in cucumber EST sequences. Similar analyses were performed in genomic and EST data from seven other plant species, and the results were compared with those of cucumber.
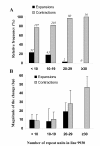
Results: A total of 112,073 perfect repeats were detected in the Gy14 cucumber genome sequence, accounting for 0.9% of the assembled Gy14 genome, with an overall density of 551.9 SSRs/Mbp. While tetranucleotides were the most frequent microsatellites in genomic DNA sequence, dinucleotide repeats, which had more repeat units than any other SSR type, had the highest cumulative sequence length. Coding regions (ESTs) of the cucumber genome had fewer microsatellites compared to its genomic sequence, with trinucleotides predominating in EST sequences. AAG was the most frequent repeat in cucumber ESTs. Overall, AT-rich motifs prevailed in both genomic and EST data. Compared to the other species examined, cucumber genomic sequence had the highest density of SSRs (although comparable to the density of poplar, grapevine and rice), and was richest in AT dinucleotides. Using an electronic PCR strategy, we investigated the polymorphism between 9930 and Gy14 at 1,006 SSR loci, and found unexpectedly high degree of polymorphism (48.3%) between the two genotypes. The level of polymorphism seems to be positively associated with the number of repeat units in the microsatellite. The in silico PCR results were validated empirically in 660 of the 1,006 SSR loci. In addition, primer sequences for more than 83,000 newly-discovered cucumber microsatellites, and their exact positions in the Gy14 genome assembly were made publicly available.
Conclusions: The cucumber genome is rich in microsatellites; AT and AAG are the most abundant repeat motifs in genomic and EST sequences of cucumber, respectively. Considering all the species investigated, some commonalities were noted, especially within the monocot and dicot groups, although the distribution of motifs and the frequency of certain repeats were characteristic of the species examined. The large number of SSR markers developed from this study should be a significant contribution to the cucurbit research community.
Figures



References
-
- Powell W, Morgante M, Andre C, Henfey M, Vogel J, Tingy S, Rafalsky A. The comparison of RFLP, RAPD, AFLP and SSR (microsatellite) markers for germplasm analysis. Mol Breed. 1996;2:225–238. doi: 10.1007/BF00564200. - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

