IL28B and the control of hepatitis C virus infection
- PMID: 20950615
- PMCID: PMC3072961
- DOI: 10.1053/j.gastro.2010.10.004
IL28B and the control of hepatitis C virus infection
Abstract
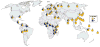
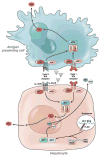
Treatment-induced control and spontaneous clearance of hepatitis C virus (HCV) infection are affected by various host factors. Polymorphisms in the region of the gene IL28B are associated with HCV clearance, implicating the gene product, interferon (IFN)-λ3, in the immune response to HCV. Although it is not clear how the IL28B haplotype affects HCV clearance, IFN-λ3 up-regulates interferon-stimulated genes, similar to IFN-α and IFN-β but via a different receptor. There is also evidence that IFN-λ3 affects the adaptive immune response. The IL28B genotype can be considered, along with other factors, in predicting patient responses to therapy with pegylated IFN-α and ribavirin. We review the genetic studies that uncovered the association between IL28B and HCV clearance, the biology of IFN-λ3, the clinical implications of the genetic association, and areas of future research.
Copyright © 2010 AGA Institute. Published by Elsevier Inc. All rights reserved.
Figures



References
-
- Wilson LE, Torbenson M, Astemborski J, Faruki H, Spoler C, Rai R, Mehta S, Kirk GD, Nelson K, Afdhal N, Thomas DL. Progression of liver fibrosis among injection drug users with chronic hepatitis C. Hepatology. 2006;43:788–795. - PubMed
-
- Thomas DL, Astemborski J, Rai RM, Anania FA, Schaeffer M, Galai N, Nolt K, Nelson KE, Strathdee SA, Johnson L, Laeyendecker O, Boitnott J, Wilson LE, Vlahov D. The natural history of hepatitis C virus infection: host, viral, and environmental factors. JAMA. 2000;284:450–456. - PubMed
-
- Thomas DL, Seeff LB. Natural history of hepatitis C. Clin Liver Dis. 2005;9:383–98. vi. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

