RETRACTED: Downregulation of p53-inducible microRNAs 192, 194, and 215 impairs the p53/MDM2 autoregulatory loop in multiple myeloma development
- PMID: 20951946
- PMCID: PMC3561766
- DOI: 10.1016/j.ccr.2010.09.005
RETRACTED: Downregulation of p53-inducible microRNAs 192, 194, and 215 impairs the p53/MDM2 autoregulatory loop in multiple myeloma development
Erratum in
-
RETRACTED: Downregulation of p53-inducible microRNAs 192, 194, and 215 Impairs the p53/MDM2 Autoregulatory Loop in Multiple Myeloma Development.Cancer Cell. 2016 Aug 8;30(2):349-351. doi: 10.1016/j.ccell.2016.07.007. Epub 2016 Aug 8. Cancer Cell. 2016. Retraction in: Cancer Cell. 2022 Nov 14;40(11):1442. doi: 10.1016/j.ccell.2022.10.007. PMID: 27505674 Retracted.
Retraction in
-
Retraction Notice to: Downregulation of p53-inducible microRNAs 192, 194, and 215 Impairs the p53/MDM2 Autoregulatory Loop in Multiple Myeloma Development.Cancer Cell. 2022 Nov 14;40(11):1441. doi: 10.1016/j.ccell.2022.10.006. Epub 2022 Oct 27. Cancer Cell. 2022. PMID: 36306793 Free PMC article. No abstract available.
Abstract
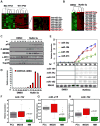
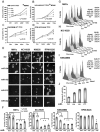
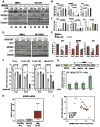
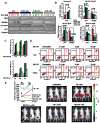
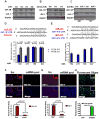
This article has been retracted: please see Elsevier Policy on Article Withdrawal (http://www.elsevier.com/locate/withdrawalpolicy). This article has been retracted at the request of the editors. This article was published on October 19, 2010, and Figures 4A and 7A were later corrected on August 8, 2016. In January 2021, The Ohio State University notified the Cancer Cell editors that an internal investigation concluded that Figures 1E, 4A, 4D, 5A, and 7A were falsified and that part of Figure 1E of the article is the same as part of Figure 1F in the correction of another article (Pichiorri et al., 2017, J. Exp. Med., 214, 1557, https://doi.org/10.1084/jem.2012095001172017c) and recommended retraction of the article. The editors no longer have confidence in the validity of the data and are retracting the article. S.-S. S. agrees with the retraction, and F.P., C.H., A.P., and C.M.C. disagree with the retraction; all other authors couldn't be reached or didn't respond.
Copyright © 2010 Elsevier Inc. All rights reserved.
Figures


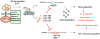
Comment in
-
Epigenetics and microRNAs combine to modulate the MDM2/p53 axis in myeloma.Cancer Cell. 2010 Oct 19;18(4):299-300. doi: 10.1016/j.ccr.2010.10.004. Cancer Cell. 2010. PMID: 20951939
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

