Substrate channeling in mammalian base excision repair pathways: passing the baton
- PMID: 20952393
- PMCID: PMC3003346
- DOI: 10.1074/jbc.M110.155267
Substrate channeling in mammalian base excision repair pathways: passing the baton
Abstract
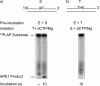
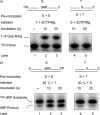
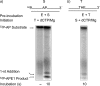
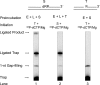
The current model for base excision repair (BER) involves two general sub-pathways termed single-nucleotide BER and long patch BER that are distinguished by their repair patch sizes and the enzymes/co-factors involved. Both sub-pathways involve a series of sequential steps from initiation to completion of repair. The BER sub-pathways are designed to sequester the various intermediates, passing them along from one step to the next without allowing these toxic molecules to trigger cell cycle arrest, necrotic cell death, or apoptosis. Although a variety of DNA-protein and protein-protein interactions are known for the BER intermediates and enzymes/co-factors, the molecular mechanisms accounting for step-to-step coordination are not well understood. In the present study we designed an in vitro assay to explore the question of whether there is a channeling or "hand-off" of the repair intermediates during BER in vitro. The results show that when BER enzymes are pre-bound to the initial single-nucleotide BER intermediate, the DNA is channeled from apurinic/apyrimidinic endonuclease 1 to DNA polymerase β and then to DNA ligase. In the long patch BER subpathway, where the 5'-end of the incised strand is blocked, the intermediate after DNA polymerase β gap filling is not channeled to the subsequent enzyme, flap endonuclease 1. Instead, flap endonuclease 1 must recognize and bind to the intermediate in competition with other molecules.
Figures





Similar articles
-
A chemical and kinetic perspective on base excision repair of DNA.Acc Chem Res. 2014 Apr 15;47(4):1238-46. doi: 10.1021/ar400275a. Epub 2014 Mar 19. Acc Chem Res. 2014. PMID: 24646203 Free PMC article.
-
A review of recent experiments on step-to-step "hand-off" of the DNA intermediates in mammalian base excision repair pathways.Mol Biol (Mosk). 2011 Jul-Aug;45(4):586-600. Mol Biol (Mosk). 2011. PMID: 21954590 Free PMC article. Review.
-
Visualizing the coordination of apurinic/apyrimidinic endonuclease (APE1) and DNA polymerase β during base excision repair.J Biol Chem. 2023 May;299(5):104636. doi: 10.1016/j.jbc.2023.104636. Epub 2023 Mar 22. J Biol Chem. 2023. PMID: 36963489 Free PMC article.
-
Coordination of steps in single-nucleotide base excision repair mediated by apurinic/apyrimidinic endonuclease 1 and DNA polymerase beta.J Biol Chem. 2007 May 4;282(18):13532-41. doi: 10.1074/jbc.M611295200. Epub 2007 Mar 12. J Biol Chem. 2007. PMID: 17355977 Free PMC article.
-
When DNA repair goes wrong: BER-generated DNA-protein crosslinks to oxidative lesions.DNA Repair (Amst). 2016 Aug;44:103-109. doi: 10.1016/j.dnarep.2016.05.014. Epub 2016 May 20. DNA Repair (Amst). 2016. PMID: 27264558 Free PMC article. Review.
Cited by
-
A chemical and kinetic perspective on base excision repair of DNA.Acc Chem Res. 2014 Apr 15;47(4):1238-46. doi: 10.1021/ar400275a. Epub 2014 Mar 19. Acc Chem Res. 2014. PMID: 24646203 Free PMC article.
-
The current state of eukaryotic DNA base damage and repair.Nucleic Acids Res. 2015 Dec 2;43(21):10083-101. doi: 10.1093/nar/gkv1136. Epub 2015 Oct 30. Nucleic Acids Res. 2015. PMID: 26519467 Free PMC article. Review.
-
Repair pathway coordination from gap filling by polβ and subsequent nick sealing by LIG1 or LIG3α governs BER efficiency at the downstream steps.DNA Repair (Amst). 2025 Apr;148:103826. doi: 10.1016/j.dnarep.2025.103826. Epub 2025 Mar 10. DNA Repair (Amst). 2025. PMID: 40081282 Free PMC article.
-
Incidence and persistence of 8-oxo-7,8-dihydroguanine within a hairpin intermediate exacerbates a toxic oxidation cycle associated with trinucleotide repeat expansion.DNA Repair (Amst). 2011 Aug 15;10(8):887-96. doi: 10.1016/j.dnarep.2011.06.003. Epub 2011 Jul 2. DNA Repair (Amst). 2011. PMID: 21727036 Free PMC article.
-
Mammalian Base Excision Repair: Functional Partnership between PARP-1 and APE1 in AP-Site Repair.PLoS One. 2015 May 28;10(5):e0124269. doi: 10.1371/journal.pone.0124269. eCollection 2015. PLoS One. 2015. PMID: 26020771 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

