Characterization of the protocatechuate 4,5-cleavage pathway operon in Comamonas sp. strain E6 and discovery of a novel pathway gene
- PMID: 20952641
- PMCID: PMC3008244
- DOI: 10.1128/AEM.01863-10
Characterization of the protocatechuate 4,5-cleavage pathway operon in Comamonas sp. strain E6 and discovery of a novel pathway gene
Abstract
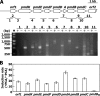
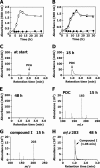
The protocatechuate (PCA) 4,5-cleavage (PCA45) pathway is the essential catabolic route for the degradation of various aromatic acids in the genus Comamonas. All of the PCA45 pathway genes, orf1-pmdKEFDABC, as well as another PCA 4,5-dioxygenase gene, pmdA(II)B(II), were isolated from a phthalate-degrading bacterium, Comamonas sp. strain E6. Disruption of pmdB and pmdD in E6, which code for the β subunit of PCA 4,5-dioxygenase and 2-pyrone-4,6-dicarboxylate (PDC) hydrolase, respectively, resulted in a growth defect on PCA, indicating that these genes are essential for the growth of E6 on PCA. On the other hand, inactivation of pmdB(II) did not affect the growth of E6 on PCA. Disruption of pmdK, which is related to a 4-hydroxybenzoate/PCA transporter of Pseudomonas putida, resulted in growth retardation on PCA. The insertional inactivation of orf1 in E6, whose deduced amino acid sequence has no similarity with proteins of known function, led to the complete loss of growth on PCA and the accumulation of PDC and 4-oxalomesaconate (OMA) from PCA. These results indicated the involvement of orf1 in the PCA45 pathway, and this gene, designated pmdU, was suggested to code for OMA tautomerase. Reverse transcription-PCR analysis suggested that the pmdUKEFDABC genes constitute an operon. The transcription start site of the pmd operon was mapped at 167 nucleotides upstream of the initiation codon of pmdU. The pmd promoter activity was enhanced 20-fold when the cells were grown in the presence of PCA. Inducers of the pmd operon were found to be PCA and PDC, but PDC was the more effective inducer.
Figures




References
-
- Bagdasarian, M., R. Lurz, B. Rückert, F. C. Franklin, M. M. Bagdasarian, J. Frey, and K. N. Timmis. 1981. Specific-purpose plasmid cloning vectors II. Broad host range, high copy number, RSF1010-derived vectors, and a host-vector system for gene cloning in Pseudomonas. Gene 16:237-247. - PubMed
-
- Blatny, J. M., T. Brautaset, H. C. Winther-Larsen, P. Karunakaran, and S. Valla. 1997. Improved broad-host-range RK2 vectors useful for high and low regulated gene expression levels in gram-negative bacteria. Plasmid 38:35-51. - PubMed
-
- Bolivar, F., and K. Backman. 1979. Plasmids of Escherichia coli as cloning vectors. Methods Enzymol. 68:245-267. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

