Construction of a single-chain variable-fragment antibody against the superantigen Staphylococcal enterotoxin B
- PMID: 20952642
- PMCID: PMC3008221
- DOI: 10.1128/AEM.01441-10
Construction of a single-chain variable-fragment antibody against the superantigen Staphylococcal enterotoxin B
Abstract
Staphylococcal food poisoning (SFP) is one of the most prevalent causes of food-borne illness throughout the world. SFP is caused by 21 different types of staphylococcal enterotoxins produced by Staphylococcus aureus. Among these, staphylococcal enterotoxin B (SEB) is the most potent toxin and is a listed biological warfare (BW) agent. Therefore, development of immunological reagents for detection of SEB is of the utmost importance. High-affinity and specific monoclonal antibodies are being used for detection of SEB, but hybridoma clones tend to lose their antibody-secreting ability over time. This problem can be overcome by the use of recombinant antibodies produced in a bacterial system. In the present investigation, genes from a hybridoma clone encoding monoclonal antibody against SEB were immortalized using antibody phage display technology. A murine phage display library containing single-chain variable-fragment (ScFv) antibody genes was constructed in a pCANTAB 5E phagemid vector. Phage particles displaying ScFv were rescued by reinfection of helper phage followed by four rounds of biopanning for selection of SEB binding ScFv antibody fragments by using phage enzyme-linked immunosorbent assay (ELISA). Soluble SEB-ScFv antibodies were characterized from one of the clones showing high affinity for SEB. The anti-SEB ScFv antibody was highly specific, and its affinity constant was 3.16 nM as determined by surface plasmon resonance (SPR). These results demonstrate that the recombinant antibody constructed by immortalizing the antibody genes from a hybridoma clone is useful for immunodetection of SEB.
Figures
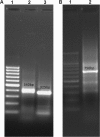



Similar articles
-
Construction of Recombinant Single Chain Variable Fragment (ScFv) Antibody Against Superantigen for Immunodetection Using Antibody Phage Display Technology.Methods Mol Biol. 2016;1396:207-225. doi: 10.1007/978-1-4939-3344-0_17. Methods Mol Biol. 2016. PMID: 26676049
-
Generation of Fab fragment-like molecular recognition proteins against staphylococcal enterotoxin B by phage display technology.Clin Vaccine Immunol. 2010 Nov;17(11):1708-17. doi: 10.1128/CVI.00229-10. Epub 2010 Sep 15. Clin Vaccine Immunol. 2010. PMID: 20844088 Free PMC article.
-
Rapid and Sensitive Detection of Staphylococcal Enterotoxin B by Recombinant Nanobody Using Phage Display Technology.Appl Biochem Biotechnol. 2019 Feb;187(2):493-505. doi: 10.1007/s12010-018-2762-y. Epub 2018 Jul 9. Appl Biochem Biotechnol. 2019. PMID: 29984392
-
Single-chain fragment variable produced by phage display technology: Construction, selection, mutation, expression, and recent applications in food safety.Compr Rev Food Sci Food Saf. 2022 Sep;21(5):4354-4377. doi: 10.1111/1541-4337.13018. Epub 2022 Jul 29. Compr Rev Food Sci Food Saf. 2022. PMID: 35904244 Review.
-
The Potential of Single-Chain Variable Fragment Antibody: Role in Future Therapeutic and Diagnostic Biologics.J Immunol Res. 2024 Aug 9;2024:1804038. doi: 10.1155/2024/1804038. eCollection 2024. J Immunol Res. 2024. PMID: 39156005 Free PMC article. Review.
Cited by
-
Identification of two neutralizing human single-chain variable fragment antibodies targeting Staphylococcus aureus alpha-hemolysin.Iran J Basic Med Sci. 2022 Oct;25(10):1207-1214. doi: 10.22038/IJBMS.2022.64103.14253. Iran J Basic Med Sci. 2022. PMID: 36311199 Free PMC article.
-
Molecular dynamics simulation of the brain-isolated single-domain antibody/nanobody from camels through in vivo phage display screening.Front Mol Biosci. 2024 Sep 2;11:1414119. doi: 10.3389/fmolb.2024.1414119. eCollection 2024. Front Mol Biosci. 2024. PMID: 39290991 Free PMC article.
-
Generation of Recombinant Antibodies Against Toxins and Viruses by Phage Display for Diagnostics and Therapy.Adv Exp Med Biol. 2016;917:55-76. doi: 10.1007/978-3-319-32805-8_4. Adv Exp Med Biol. 2016. PMID: 27236552 Free PMC article. Review.
-
Construction and characterization of VL-VH tail-parallel genetically engineered antibodies against staphylococcal enterotoxins.Immunol Res. 2015 Mar;61(3):281-93. doi: 10.1007/s12026-015-8623-7. Immunol Res. 2015. PMID: 25608796
-
Chronocoulometric aptamer based assay for staphylococcal enterotoxin B by target-triggered assembly of nanostructured dendritic nucleic acids on a gold electrode.Mikrochim Acta. 2019 Jan 14;186(2):109. doi: 10.1007/s00604-019-3236-9. Mikrochim Acta. 2019. PMID: 30637509
References
-
- Altschul, S. F., W. Gish, W. Miller, E. W. Mayers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. - PubMed
-
- Azzazy, H. M. E., and W. E. Highsmith, Jr. 2002. Phage display technology: clinical applications and recent innovations. Clin. Biochem. 35:425-445. - PubMed
-
- Baker, M. D., A. C. Papageorgiou, R. W. Titball, J. Miller, S. White, B. Lingard, J. J. Lee, D. Cavanagh, M. A. Kehoe, J. H. Robinson, and K. R. Acharya. 2002. Structural and functional role of threonine 112 in a superantigen Staphylococcus aureus enterotoxin B. J. Biol. Chem. 277:2756-2762. - PubMed
-
- Balaban, N., and A. Rasooly. 2000. Staphylococcal enterotoxins. Int. J. Food Microbiol. 61:1-10. - PubMed
-
- Bergdoll, M. S. 1983. Enterotoxins, p. 559-598. In C. S. F. Easmon and C. Adlam (ed.), Staphylococci and staphylococcal infections. Academic Press, New York, NY.
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources

