Activation of a silent fungal polyketide biosynthesis pathway through regulatory cross talk with a cryptic nonribosomal peptide synthetase gene cluster
- PMID: 20952652
- PMCID: PMC3008269
- DOI: 10.1128/AEM.00683-10
Activation of a silent fungal polyketide biosynthesis pathway through regulatory cross talk with a cryptic nonribosomal peptide synthetase gene cluster
Abstract
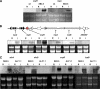
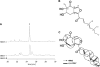
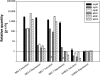
Filamentous fungi produce numerous natural products that constitute a consistent source of potential drug leads, yet it seems that the majority of natural products are overlooked since most biosynthesis gene clusters are silent under standard cultivation conditions. Screening secondary metabolite genes of the model fungus Aspergillus nidulans, we noted a silent gene cluster on chromosome II comprising two nonribosomal peptide synthetase (NRPS) genes, inpA and inpB, flanked by a regulatory gene that we named scpR for secondary metabolism cross-pathway regulator. The induced expression of the scpR gene using the promoter of the alcohol dehydrogenase AlcA led to the transcriptional activation of both the endogenous scpR gene and the NRPS genes. Surprisingly, metabolic profiling of the supernatant of mycelia overexpressing scpR revealed the production of the polyketide asperfuranone. Through transcriptome analysis we found that another silent secondary metabolite gene cluster located on chromosome VIII coding for asperfuranone biosynthesis was specifically induced. Quantitative reverse transcription-PCR proved the transcription not only of the corresponding polyketide synthase (PKS) biosynthesis genes, afoE and afoG, but also of their activator, afoA, under alcAp-scpR-inducing conditions. To exclude the possibility that the product of the inp cluster induced the asperfuranone gene cluster, a strain carrying a deletion of the NRPS gene inpB and, in addition, the alcAp-scpR overexpression cassette was generated. In this strain, under inducing conditions, transcripts of the biosynthesis genes of both the NRPS-containing gene cluster inp and the asperfuranone gene cluster except gene inpB were detected. Moreover, the existence of the polyketide product asperfuranone indicates that the transcription factor ScpR controls the expression of the asperfuranone biosynthesis gene cluster. This expression as well as the biosynthesis of asperfuranone was abolished after the deletion of the asperfuranone activator gene afoA, indicating that ScpR binds to the afoA promoter. To the best of our knowledge, this is the first report of regulatory cross talk between two biosynthesis gene clusters located on different chromosomes.
Figures

References
-
- Bailey, T. L., and C. Elkan. 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 2:28-36. - PubMed
-
- Bergmann, S., J. Schuemann, K. Scherlach, C. Lange, A. A. Brakhage, and C. Hertweck. 2007. Genomics-driven discovery of PKS-NRPS hybrid metabolites from Aspergillus nidulans. Nat. Chem. Biol. 3:213-217. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

