Structural basis for cooperative RNA binding and export complex assembly by HIV Rev
- PMID: 20953181
- PMCID: PMC2988976
- DOI: 10.1038/nsmb.1902
Structural basis for cooperative RNA binding and export complex assembly by HIV Rev
Abstract
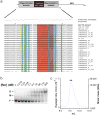
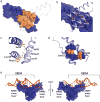
HIV replication requires nuclear export of unspliced viral RNAs to translate structural proteins and package genomic RNA. Export is mediated by cooperative binding of the Rev protein to the Rev response element (RRE) RNA, to form a highly specific oligomeric ribonucleoprotein (RNP) that binds to the Crm1 host export factor. To understand how protein oligomerization generates cooperativity and specificity for RRE binding, we solved the crystal structure of a Rev dimer at 2.5-Å resolution. The dimer arrangement organizes arginine-rich helices at the ends of a V-shaped assembly to bind adjacent RNA sites and structurally couple dimerization and RNA recognition. A second protein-protein interface arranges higher-order Rev oligomers to act as an adaptor to the host export machinery, with viral RNA bound to one face and Crm1 to another, the oligomers thereby using small, interconnected modules to physically arrange the RNP for efficient export.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Cullen BR. Nuclear mRNA export: insights from virology. Trends Biochem Sci. 2003;28:419–24. - PubMed
-
- Pollard VW, Malim MH. The HIV-1 Rev Protein. Annu Rev Microbiol. 1998;52:491–532. - PubMed
-
- Fornerod M, Ohno M, Yoshida M, Mattaj IW. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell. 1997;90:1051–1060. - PubMed
-
- Malim MH, Cullen BR. HIV-1 structural gene expression requires the binding of multiple Rev monomers to the viral RRE: implications for HIV-1 latency. Cell. 1991;65:241–48. - PubMed
-
- Mann DA, et al. A molecular rheostat: Co-operative rev binding to stem I of the Rev-response element modulates human immunodeficiency virus type-1 late gene expression. J Mol Biol. 1994;241:193–207. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

