Marine genomics: at the interface of marine microbial ecology and biodiscovery
- PMID: 20953417
- PMCID: PMC2948669
- DOI: 10.1111/j.1751-7915.2010.00193.x
Marine genomics: at the interface of marine microbial ecology and biodiscovery
Abstract
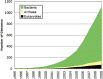
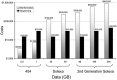
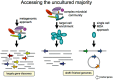
The composition and activities of microbes from diverse habitats have been the focus of intense research during the past decade with this research being spurred on largely by advances in molecular biology and genomic technologies. In recent years environmental microbiology has entered very firmly into the age of the 'omics' – (meta)genomics, proteomics, metabolomics, transcriptomics – with probably others on the rise. Microbes are essential participants in all biogeochemical processes on our planet, and the practical applications of what we are learning from the use of molecular approaches has altered how we view biological systems. In addition, there is considerable potential to use information about uncultured microbes in biodiscovery research as microbes provide a rich source of discovery for novel genes, enzymes and metabolic pathways. This review explores the brief history of genomic and metagenomic approaches to study environmental microbial assemblages and describes some of the future challenges involved in broadening our approaches – leading to new insights for understanding environmental problems and enabling biodiscovery research.
Figures
Similar articles
-
Marine genomics: at the interface of marine microbial ecology and biotechnological applications. Proceedings of a CIESM-NSF/EC Workshop. October 12-14, 2008. Monaco.Microb Biotechnol. 2010 Sep;3(5):489-621. Microb Biotechnol. 2010. PMID: 21534258 No abstract available.
-
Genomic approaches in marine biodiversity and aquaculture.Biol Res. 2013;46(4):353-61. doi: 10.4067/S0716-97602013000400007. Biol Res. 2013. PMID: 24510138
-
Marine Protists Are Not Just Big Bacteria.Curr Biol. 2017 Jun 5;27(11):R541-R549. doi: 10.1016/j.cub.2017.03.075. Curr Biol. 2017. PMID: 28586691 Review.
-
Marine microbial genomics in Europe: current status and perspectives.Microb Biotechnol. 2010 Sep;3(5):523-30. doi: 10.1111/j.1751-7915.2010.00169.x. Microb Biotechnol. 2010. PMID: 20953416 Free PMC article. Review.
-
Construction and Screening of Marine Metagenomic Large Insert Libraries.Methods Mol Biol. 2017;1539:23-42. doi: 10.1007/978-1-4939-6691-2_3. Methods Mol Biol. 2017. PMID: 27900682
Cited by
-
Antioxidant Molecules from Marine Fungi: Methodologies and Perspectives.Antioxidants (Basel). 2020 Nov 26;9(12):1183. doi: 10.3390/antiox9121183. Antioxidants (Basel). 2020. PMID: 33256101 Free PMC article. Review.
-
Whole cell biocatalysts: essential workers from Nature to the industry.Microb Biotechnol. 2017 Mar;10(2):250-263. doi: 10.1111/1751-7915.12363. Epub 2016 May 3. Microb Biotechnol. 2017. PMID: 27145540 Free PMC article. Review.
-
The metagenome of an anaerobic microbial community decomposing poplar wood chips.PLoS One. 2012;7(5):e36740. doi: 10.1371/journal.pone.0036740. Epub 2012 May 21. PLoS One. 2012. PMID: 22629327 Free PMC article.
-
Analysis of metagenomic data containing high biodiversity levels.PLoS One. 2013;8(3):e58118. doi: 10.1371/journal.pone.0058118. Epub 2013 Mar 7. PLoS One. 2013. PMID: 23505458 Free PMC article.
-
Characterization of the Jomthonic Acids Biosynthesis Pathway and Isolation of Novel Analogues in Streptomyces caniferus GUA-06-05-006A.Mar Drugs. 2018 Jul 31;16(8):259. doi: 10.3390/md16080259. Mar Drugs. 2018. PMID: 30065171 Free PMC article.
References
-
- Banfield J.F., Young M. Microbiology. Variety the spice of life – in microbial communities. Science. 2009;326:1198–1199. - PubMed
-
- Béjà O., Suzuki M.T., Koonin E.V., Aravind L., Hadd A., Nguyen L.P. Construction and analysis of bacterial artificial chromosome libraries from a marine microbial assemblage. Environ Microbiol. 2000a;2:516–529. et al. - PubMed
-
- Béjà O., Aravind L., Koonin E.V., Suzuki M.T., Hadd A. Bacterial Rhodopsin: Evidence for a new type of phototrophy in the sea. Science. 2000b;289:1902–1906. et al. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

