Transforming growth factor-β-inducible early response gene 1 is a novel substrate for atypical protein kinase Cs
- PMID: 20953893
- PMCID: PMC3092057
- DOI: 10.1007/s00018-010-0541-1
Transforming growth factor-β-inducible early response gene 1 is a novel substrate for atypical protein kinase Cs
Abstract
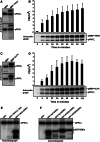
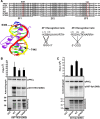
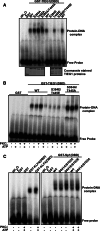
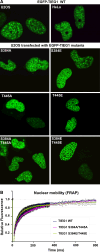
The protein kinase C (PKC) family of serine/threonine kinases consists of ten different isoforms grouped into three subfamilies, denoted classical, novel and atypical PKCs (aPKCs). The aPKCs, PKCι/λ and PKCζ serve important roles during development and in processes subverted in cancer such as cell and tissue polarity, cell proliferation, differentiation and apoptosis. In an effort to identify novel interaction partners for aPKCs, we performed a yeast two-hybrid screen with the regulatory domain of PKCι/λ as bait and identified the Krüppel-like factors family protein TIEG1 as a putative interaction partner for PKCι/λ. We confirmed the interaction of both aPKCs with TIEG1 in vitro and in cells, and found that both aPKCs phosphorylate the DNA-binding domain of TIEG1 on two critical residues. Interestingly, the aPKC-mediated phosphorylation of TIEG1 affected its DNA-binding activity, subnuclear localization and transactivation potential.
Figures


Similar articles
-
The human papillomavirus-16 (HPV-16) oncoprotein E7 conjugates with and mediates the role of the transforming growth factor-beta inducible early gene 1 (TIEG1) in apoptosis.Int J Biochem Cell Biol. 2010 Nov;42(11):1831-9. doi: 10.1016/j.biocel.2010.07.019. Epub 2010 Aug 6. Int J Biochem Cell Biol. 2010. PMID: 20691807
-
Rab2 interacts directly with atypical protein kinase C (aPKC) iota/lambda and inhibits aPKCiota/lambda-dependent glyceraldehyde-3-phosphate dehydrogenase phosphorylation.J Biol Chem. 2003 Dec 26;278(52):52524-30. doi: 10.1074/jbc.M309343200. Epub 2003 Oct 21. J Biol Chem. 2003. PMID: 14570876
-
Crystal structure of the catalytic domain of human atypical protein kinase C-iota reveals interaction mode of phosphorylation site in turn motif.J Mol Biol. 2005 Sep 30;352(4):918-31. doi: 10.1016/j.jmb.2005.07.060. J Mol Biol. 2005. PMID: 16125198
-
Role of TIEG1 in biological processes and disease states.J Cell Biochem. 2007 Oct 15;102(3):539-48. doi: 10.1002/jcb.21492. J Cell Biochem. 2007. PMID: 17729309 Review.
-
Targeting the oncogenic protein kinase Ciota signalling pathway for the treatment of cancer.Biochem Soc Trans. 2007 Nov;35(Pt 5):996-1000. doi: 10.1042/BST0350996. Biochem Soc Trans. 2007. PMID: 17956262 Review.
Cited by
-
The lncRNA SLNCR Recruits the Androgen Receptor to EGR1-Bound Genes in Melanoma and Inhibits Expression of Tumor Suppressor p21.Cell Rep. 2019 May 21;27(8):2493-2507.e4. doi: 10.1016/j.celrep.2019.04.101. Cell Rep. 2019. PMID: 31116991 Free PMC article.
-
Differential coupling of KLF10 to Sin3-HDAC and PCAF regulates the inducibility of the FOXP3 gene.Am J Physiol Regul Integr Comp Physiol. 2014 Sep 15;307(6):R608-20. doi: 10.1152/ajpregu.00085.2014. Epub 2014 Jun 18. Am J Physiol Regul Integr Comp Physiol. 2014. PMID: 24944246 Free PMC article.
-
TIEG1 modulates β-catenin sub-cellular localization and enhances Wnt signaling in bone.Nucleic Acids Res. 2017 May 19;45(9):5170-5182. doi: 10.1093/nar/gkx118. Nucleic Acids Res. 2017. PMID: 28201653 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

