Cotton plants expressing CYP6AE14 double-stranded RNA show enhanced resistance to bollworms
- PMID: 20953975
- PMCID: PMC3090577
- DOI: 10.1007/s11248-010-9450-1
Cotton plants expressing CYP6AE14 double-stranded RNA show enhanced resistance to bollworms
Abstract
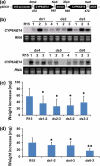
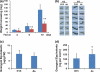
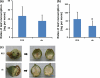
RNA interference (RNAi) plays an important role in regulating gene expression in eukaryotes. Previously, we generated Arabidopsis and tobacco plants expressing double-stranded RNA (dsRNA) targeting a cotton bollworm (Helicoverpa armigera) P450 gene, CYP6AE14. Bollworms fed on transgenic dsCYP6AE14 plants showed suppressed CYP6AE14 expression and reduced growth on gossypol-containing diet (Mao et al., in Nat Biotechnol 25: 1307-1313, 2007). Here we report generation and analysis of dsRNA-expressing cotton (Gossypium hirsutum) plants. Bollworm larvae reared on T2 plants of the ds6-3 line exhibited drastically retarded growth, and the transgenic plants were less damaged by bollworms than the control. Quantitative reverse-transcription polymerase chain reaction (RT-PCR) showed that the CYP6AE14 expression level was reduced in the larvae as early as 4 h after feeding on the transgenic plants; accordingly, the CYP6AE14 protein level dropped. These results demonstrated that transgenic cotton plants expressing dsCYP6AE14 acquired enhanced resistance to cotton bollworms, and that RNAi technology can be used for engineering insect-proof cotton cultivar.
Figures


Similar articles
-
Cysteine protease enhances plant-mediated bollworm RNA interference.Plant Mol Biol. 2013 Sep;83(1-2):119-29. doi: 10.1007/s11103-013-0030-7. Epub 2013 Mar 4. Plant Mol Biol. 2013. PMID: 23460027 Free PMC article.
-
Silencing a cotton bollworm P450 monooxygenase gene by plant-mediated RNAi impairs larval tolerance of gossypol.Nat Biotechnol. 2007 Nov;25(11):1307-13. doi: 10.1038/nbt1352. Epub 2007 Nov 4. Nat Biotechnol. 2007. PMID: 17982444
-
Transgenic Cotton Plants Expressing Double-stranded RNAs Target HMG-CoA Reductase (HMGR) Gene Inhibits the Growth, Development and Survival of Cotton Bollworms.Int J Biol Sci. 2015 Sep 15;11(11):1296-305. doi: 10.7150/ijbs.12463. eCollection 2015. Int J Biol Sci. 2015. PMID: 26435695 Free PMC article.
-
Impacts of Bt transgenic cotton on integrated pest management.J Agric Food Chem. 2011 Jun 8;59(11):5842-51. doi: 10.1021/jf102939c. Epub 2010 Oct 13. J Agric Food Chem. 2011. PMID: 20942488 Review.
-
Diet-delivered RNAi in Helicoverpa armigera--Progresses and challenges.J Insect Physiol. 2016 Feb;85:86-93. doi: 10.1016/j.jinsphys.2015.11.005. Epub 2015 Nov 5. J Insect Physiol. 2016. PMID: 26549127 Review.
Cited by
-
Transcriptome Analysis and Screening for Potential Target Genes for RNAi-Mediated Pest Control of the Beet Armyworm, Spodoptera exigua.PLoS One. 2013 Jun 18;8(6):e65931. doi: 10.1371/journal.pone.0065931. Print 2013. PLoS One. 2013. PMID: 23823756 Free PMC article.
-
Knock-Down of Gossypol-Inducing Cytochrome P450 Genes Reduced Deltamethrin Sensitivity in Spodoptera exigua (Hübner).Int J Mol Sci. 2019 May 7;20(9):2248. doi: 10.3390/ijms20092248. Int J Mol Sci. 2019. PMID: 31067723 Free PMC article.
-
RNAi technology: a new platform for crop pest control.Physiol Mol Biol Plants. 2017 Jul;23(3):487-501. doi: 10.1007/s12298-017-0443-x. Epub 2017 Apr 29. Physiol Mol Biol Plants. 2017. PMID: 28878489 Free PMC article. Review.
-
RNA-Seq Study of Microbially Induced Hemocyte Transcripts from Larval Heliothis virescens (Lepidoptera: Noctuidae).Insects. 2012 Aug 14;3(3):743-62. doi: 10.3390/insects3030743. Insects. 2012. PMID: 26466627 Free PMC article.
-
HIGS-mediated crop protection against cotton aphids.Plant Biotechnol J. 2025 Mar;23(3):692-694. doi: 10.1111/pbi.14529. Epub 2024 Dec 5. Plant Biotechnol J. 2025. PMID: 39636291 Free PMC article. No abstract available.
References
-
- Benbouza H, Baudoin J-P, Mergeai G. Improvement of the genomic DNA extraction method with CTAB for cotton leaves. Biotechnologie Agronomie Societe et Environ. 2006;10:73–76.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

