Dual-channel single-molecule fluorescence resonance energy transfer to establish distance parameters for RNA nanoparticles
- PMID: 20954698
- PMCID: PMC2990273
- DOI: 10.1021/nn1014853
Dual-channel single-molecule fluorescence resonance energy transfer to establish distance parameters for RNA nanoparticles
Abstract
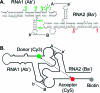
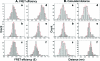
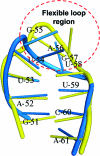
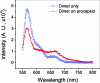
The increasing interest in RNA nanotechnology and the demonstrated feasibility of using RNA nanoparticles as therapeutics have prompted the need for imaging systems with nanometer-scale resolution for RNA studies. Phi29 dimeric pRNAs can serve as building blocks in assembly into the hexameric ring of the nanomotors, as modules of RNA nanoparciles, and as vehicles for specific delivery of therapeutics to cancers or viral infected cells. The understanding of the 3D structure of this novel RNA dimeric particle is fundamentally and practically important. Although a 3D model of pRNA dimer has been proposed based on biochemical analysis, no distance measurements or X-ray diffraction data have been reported. Here we evaluated the application of our customized single-molecule dual-viewing system for distance measurement within pRNA dimers using single-molecule Fluorescence Resonance Energy Transfer (smFRET). Ten pRNA monomers labeled with single donor or acceptor fluorophores at various locations were constructed and eight dimers were assembled. smFRET signals were detected for six dimers. The tethered arm sizes of the fluorophores were estimated empirically from dual-labeled RNA/DNA standards. The distances between donor and acceptor were calculated and used as distance parameters to assess and refine the previously reported 3D model of the pRNA dimer. Distances between nucleotides in pRNA dimers were found to be different from those of the dimers bound to procapsid, suggesting a conformational change of the pRNA dimer upon binding to the procapsid.
Figures





Similar articles
-
Crystal structure of 3WJ core revealing divalent ion-promoted thermostability and assembly of the Phi29 hexameric motor pRNA.RNA. 2013 Sep;19(9):1226-37. doi: 10.1261/rna.037077.112. Epub 2013 Jul 24. RNA. 2013. PMID: 23884902 Free PMC article.
-
Mapping the inter-RNA interaction of bacterial virus phi29 packaging RNA by site-specific photoaffinity cross-linking.J Biol Chem. 2000 Jan 28;275(4):2817-24. doi: 10.1074/jbc.275.4.2817. J Biol Chem. 2000. PMID: 10644747
-
Chemical modification patterns of active and inactive as well as procapsid-bound and unbound DNA-packaging RNAof bacterial virus Phi29.Virology. 2001 Mar 15;281(2):281-93. doi: 10.1006/viro.2000.0771. Virology. 2001. PMID: 11277700
-
RNA nanotechnology: engineering, assembly and applications in detection, gene delivery and therapy.J Nanosci Nanotechnol. 2005 Dec;5(12):1964-82. doi: 10.1166/jnn.2005.446. J Nanosci Nanotechnol. 2005. PMID: 16430131 Free PMC article. Review.
-
Structure and function of phi29 hexameric RNA that drives the viral DNA packaging motor: review.Prog Nucleic Acid Res Mol Biol. 2002;72:415-72. doi: 10.1016/s0079-6603(02)72076-x. Prog Nucleic Acid Res Mol Biol. 2002. PMID: 12206459 Review.
Cited by
-
Optimized method for the synthesis and purification of adenosine--folic acid conjugates for use as transcription initiators in the preparation of modified RNA.Methods. 2011 Jun;54(2):260-6. doi: 10.1016/j.ymeth.2010.12.007. Epub 2010 Dec 14. Methods. 2011. PMID: 21163352 Free PMC article.
-
Crystal structure of 3WJ core revealing divalent ion-promoted thermostability and assembly of the Phi29 hexameric motor pRNA.RNA. 2013 Sep;19(9):1226-37. doi: 10.1261/rna.037077.112. Epub 2013 Jul 24. RNA. 2013. PMID: 23884902 Free PMC article.
-
Global structure of a three-way junction in a phi29 packaging RNA dimer determined using site-directed spin labeling.J Am Chem Soc. 2012 Feb 8;134(5):2644-52. doi: 10.1021/ja2093647. Epub 2012 Jan 27. J Am Chem Soc. 2012. PMID: 22229766 Free PMC article.
-
Arrowtail RNA for Ligand Display on Ginger Exosome-like Nanovesicles to Systemic Deliver siRNA for Cancer Suppression.Sci Rep. 2018 Oct 2;8(1):14644. doi: 10.1038/s41598-018-32953-7. Sci Rep. 2018. PMID: 30279553 Free PMC article.
-
Single-Molecule FRET-Based Dynamic DNA Sensor.ACS Sens. 2021 Mar 26;6(3):1367-1374. doi: 10.1021/acssensors.1c00002. Epub 2021 Mar 15. ACS Sens. 2021. PMID: 33720708 Free PMC article.
References
-
- Weiss S. Fluorescence Spectroscopy of Single Biomolecules. Science 1999, 283, 1676–1683. - PubMed
-
- Leake M. C.; Chandler J. H.; Wadhams G. H.; Bai F.; Berry R. M.; Armitage J. P. Stoichiometry and Turnover in Single, Functioning Membrane Protein Complexes. Nature 2006, 443, 355–358. - PubMed
-
- Das S. K.; Darshi M.; Cheley S.; Wallace M. I.; Bayley H. Membrane Protein Stoichiometry Determined From the Step-Wise Photobleaching of Dye-Labelled Subunits. ChemBioChem 2007, 8, 994–999. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

