Epigenetic methodologies for behavioral scientists
- PMID: 20955712
- PMCID: PMC3093106
- DOI: 10.1016/j.yhbeh.2010.10.007
Epigenetic methodologies for behavioral scientists
Abstract
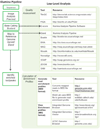
Hormones are essential regulators of many behaviors. Steroids bind either to nuclear or membrane receptors while peptides primarily act via membrane receptors. After a ligand binds, the conformational change in the receptor initiates changes in cell signaling cascades (membrane receptors) or direct alternations in DNA transcription (steroid receptors). Changes in gene transcription that result are responsible for protein production and ultimately behavioral modifications. A significant part of how hormones affect DNA transcription is via epigenetic modifications of DNA and/or the chromatin in which it is entwined. These alterations lead to transcriptional changes that ultimately define the phenotype and function of a given cell. Importantly we now know that environmental stimuli influence epigenetic marks, which in the context of neuroendocrinology can lead to behavioral changes. Importantly tracking epigenetic states and profiling the epigenome within cells require the use of epigenetic methodologies and subsequent data analysis. Here we describe the techniques of particular importance in the mapping of DNA methylation, histone modifications and occupancy of chromatin bound effector proteins that regulate gene expression. For researchers wanting to move into these levels of analysis we discuss the application of modern sequencing technologies applied in assays such as chromatin immunoprecipitation and the bioinformatics analysis involved in the rich datasets generated.
Copyright © 2010 Elsevier Inc. All rights reserved.
Figures

Similar articles
-
Epigenomics in stress tolerance of plants under the climate change.Mol Biol Rep. 2023 Jul;50(7):6201-6216. doi: 10.1007/s11033-023-08539-6. Epub 2023 Jun 9. Mol Biol Rep. 2023. PMID: 37294468 Review.
-
Investigation of epigenetics in kidney cell biology.Methods Cell Biol. 2019;153:255-278. doi: 10.1016/bs.mcb.2019.04.015. Epub 2019 Jun 13. Methods Cell Biol. 2019. PMID: 31395383 Free PMC article. Review.
-
Chromatin Profiling Techniques: Exploring the Chromatin Environment and Its Contributions to Complex Traits.Int J Mol Sci. 2021 Jul 16;22(14):7612. doi: 10.3390/ijms22147612. Int J Mol Sci. 2021. PMID: 34299232 Free PMC article. Review.
-
Profiling the Epigenetic Landscape of the Spermatogonial Stem Cell-Part 1: Epigenomics Assays.Methods Mol Biol. 2023;2656:71-108. doi: 10.1007/978-1-0716-3139-3_5. Methods Mol Biol. 2023. PMID: 37249867
-
Computational methods and next-generation sequencing approaches to analyze epigenetics data: Profiling of methods and applications.Methods. 2021 Mar;187:92-103. doi: 10.1016/j.ymeth.2020.09.008. Epub 2020 Sep 14. Methods. 2021. PMID: 32941995 Free PMC article. Review.
Cited by
-
DNA methylation: an epigenetic risk factor in preterm birth.Reprod Sci. 2012 Jan;19(1):6-13. doi: 10.1177/1933719111424446. Reprod Sci. 2012. PMID: 22228737 Free PMC article. Review.
-
Aging effects on DNA methylation modules in human brain and blood tissue.Genome Biol. 2012 Oct 3;13(10):R97. doi: 10.1186/gb-2012-13-10-r97. Genome Biol. 2012. PMID: 23034122 Free PMC article.
-
A Molecular Perspective on Procedures and Outcomes with Assisted Reproductive Technologies.Cold Spring Harb Perspect Med. 2016 Apr 1;6(4):a023416. doi: 10.1101/cshperspect.a023416. Cold Spring Harb Perspect Med. 2016. PMID: 26747835 Free PMC article. Review.
-
Neuroplasticity in addiction: cellular and transcriptional perspectives.Front Mol Neurosci. 2012 Nov 12;5:99. doi: 10.3389/fnmol.2012.00099. eCollection 2012. Front Mol Neurosci. 2012. PMID: 23162427 Free PMC article.
-
High cortisol in 5-year-old children causes loss of DNA methylation in SINE retrotransposons: a possible role for ZNF263 in stress-related diseases.Clin Epigenetics. 2015 Sep 4;7(1):91. doi: 10.1186/s13148-015-0123-z. eCollection 2015. Clin Epigenetics. 2015. PMID: 26339299 Free PMC article.
References
-
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. - PubMed
-
- Bernstein BE, Kamal M, Lindblad-Toh K, Bekiranov S, Bailey DK, Huebert DJ, McMahon S, Karlsson EK, Kulbokas EJ, 3rd, Gingeras TR, Schreiber SL, Lander ES. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell. 2005;120:169–181. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

