Multiple global suppressors of protein stability defects facilitate the evolution of extended-spectrum TEM β-lactamases
- PMID: 20955714
- PMCID: PMC3032993
- DOI: 10.1016/j.jmb.2010.10.008
Multiple global suppressors of protein stability defects facilitate the evolution of extended-spectrum TEM β-lactamases
Abstract
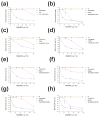
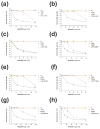
The introduction of extended-spectrum cephalosporins and β-lactamase inhibitors has driven the evolution of extended-spectrum β-lactamases (ESBLs) that possess the ability to hydrolyze these drugs. The evolved TEM ESBLs from clinical isolates of bacteria often contain substitutions that occur in the active site and alter the catalytic properties of the enzyme to provide an increased hydrolysis of extended-spectrum cephalosporins or an increased resistance to inhibitors. These active-site substitutions often result in a cost in the form of reduced enzyme stability. The evolution of TEM ESBLs is facilitated by mutations that act as global suppressors of protein stability defects in that they allow the enzyme to absorb multiple amino acid changes despite incremental losses in stability associated with the substitutions. The best-studied example is the M182T substitution, which corrects protein stability defects and is commonly found in TEM ESBLs or inhibitor-resistant variants from clinical isolates. In this study, a genetic selection for second-site mutations that could partially restore function to a severely destabilized primary mutant enabled the identification of A184V, T265M, R275Q, and N276D, which are known to occur in TEM ESBLs from clinical isolates, as suppressors of TEM-1 protein stability defects. Further characterization demonstrated that these substitutions increased the thermal stability of TEM-1 and were able to correct the stability defects of two different sets of destabilizing mutations. The acquisition of compensatory global suppressors of stability costs associated with active-site mutations may be a common mechanism for the evolution of novel protein function.
Copyright © 2010 Elsevier Ltd. All rights reserved.
Figures




References
-
- Orencia MC, Yoon JS, Ness JE, Stemmer WP, Stevens RC. Predicting the emergence of antibiotic resistance by directed evolution and structural analysis. Nat Struct Biol. 2001;8:238–242. - PubMed
-
- Wang X, Minasov G, Shoichet BK. Evolution of an antibiotic resistance enzyme constrained by stability and activity trade-offs. J Mol Biol. 2002;320:85–95. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

